Research Topics:
Molecular Mechanisms of Collagen Homeostasis
Collagens are the most abundant animal proteins found as main constituents of connective tissue and several organs, as well as key weaving components of the extracellular matrix (ECM). In humans, more than 50 different genes encode for cross-linked collagen polypeptides, that assemble into 28 homo- and heteromeric triple helical assemblies characterized by Gly-X-Y sequence repeats. Collagen molecules undergo extensive post-translational modification (PTM) in the endoplasmic reticulum (ER), enabling triple-helix formation and subsequent extracellular assembly into fibers, mesh networks and other oligomers, depending on the collagen type. Our group has been focusing on these enzymes for many years.
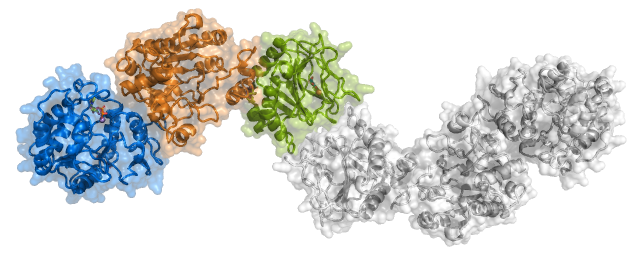
We have determined the first crystal structure of a full-length human Procollagen Lysyl 2-Oxoglutarate Dioxygenase LH enzyme (LH3/PLOD3), providing a structural framework to understand the molecular mechanisms of collagen lysine modification and to rationalize the impact of the numerous disease-related mutations affecting PLOD genes. Our research has then expanded on biochemical and biophysical studies of LH variants and their roles in homeostasis and disease.
|
Crystal structure of LH3/PLOD3. Shown is the elongated dimeric molecular architecture of the enzyme, characterized by three domains composing each monomer named (from the N-terminus) GT (Glycosyltransferase), AC (i.e., Accessory), and LH (i.e., Lysyl Hydroxylase). |
Aiming at rationalizing the complete Lysine-to-GlucosylGalactosylHydroxylysine pathway, we focused on hydroxylysine galactosyltransferase GLT25D1/COLGALT1. The crystal structure of this enzyme revealed, similar to LH3/PLOD3, an elongated dimeric architecture. Each monomer hosts two Rossman fold type domains. Surprisingly, both domains are populated with donor substrates and metal ions, but only the external C-terminal GT2 domain has catalytic activity and binds Mn2+. The GT1 domain, shaping the enzyme’s dimer interface, binds Ca2+ and UDP-Galactose for structural purposes and shapes the interface required for the interaction between GLT25D1/COLGALT1 and LH3/PLOD3.

|
Crystal structure of GLT25D1/COLGALT1. Shown is the elongated dimeric molecular architecture of the enzyme, characterized by Two domains composing each monomer named (from the N-terminus) GT1 (structural, Ca2+ binding), , and GT2 (catalytic, Mn2+ binding). |
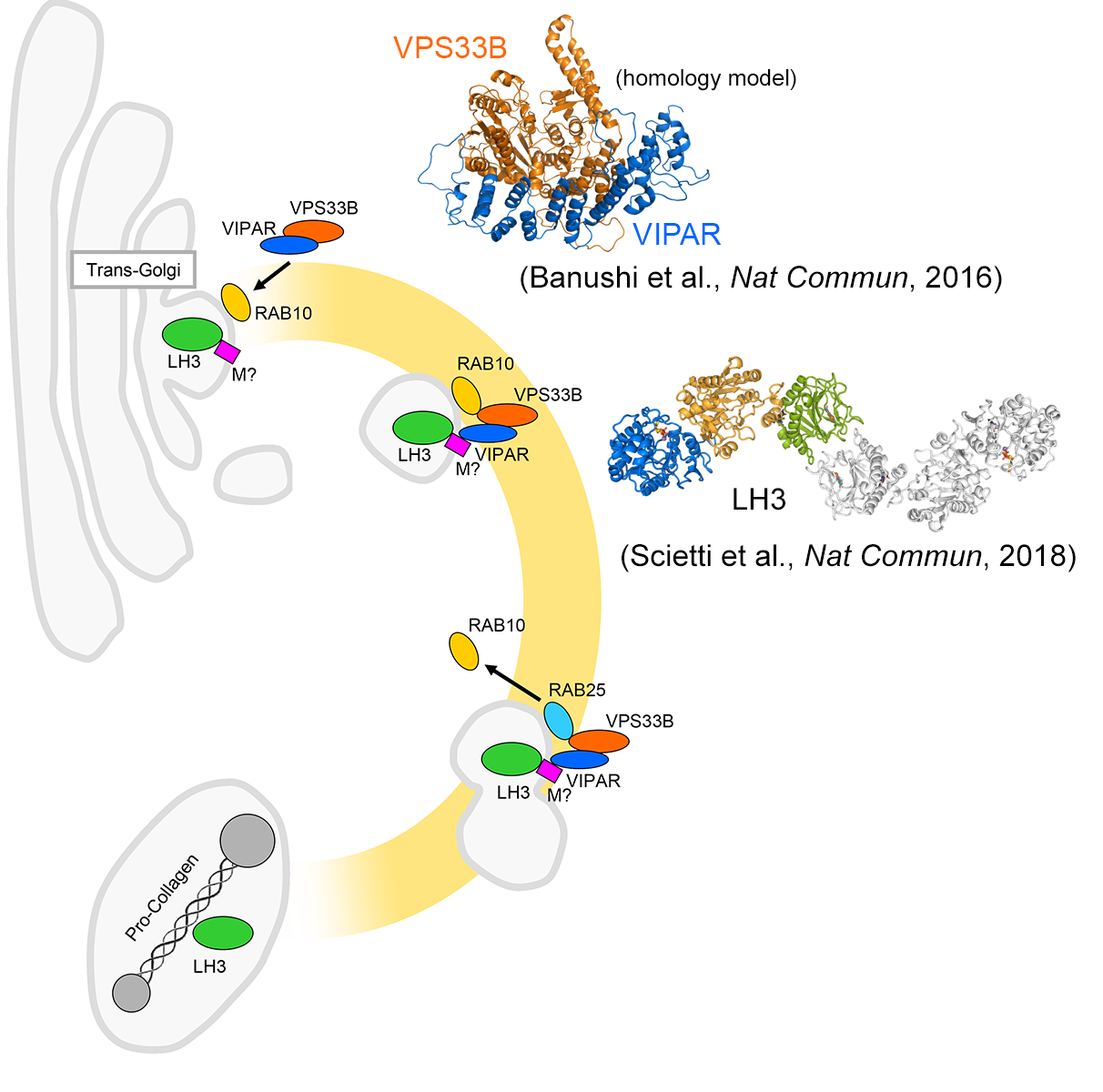
Earlier in 2016, in collaboration with the group of Prof. Paul Gissen at the UCL London, we discovered that VPS33B/VIPAR-mediated trafficking of LH3 to newly described cytoplasmic organelles (collagen IV carriers or CIVC) is essential to post-translational modification of de novo generated collagen, its structure and function. This represents the discovery of a novel post-Golgi trafficking pathway that involves previously unrelated sorting proteins and enzymes, also defining roles for the previously uncharacterized VPS33B and VIPAR proteins in regulation of LH3/PLOD3 enzyme functions. VPS33B and VIPAR are deficient in the severe multisystem disorder Arthrogryposis, Renal dysfunction and Cholestasis syndrome (ARC). Our data provide new insights to understand the molecular mechanisms of this devastating genetic syndrome and related phenotypes.
|
Schematic of the pathway targeting LH3 from the trans-Golgi Network to the newly described procollagen containing organelles is regulated by VIPAR and its interacting proteins. Shown on the top-right corner is the homology model of VPS33B-VIPAR interaction, on the right is the crystal structre of full-lenght, dimeric human LH3. |
Software:
PDB Files:
SASBDB Files:
Files in Repository:
Publications:
Molecular Structure and Enzymatic Mechanism of the Human Collagen Hydroxylysine Galactosyltransferase GLT25D1/COLGALT1
Nature Communications, 16, 3624. (2025) *Co-first authors - PubMed
Identification of regulatory molecular "hot spots" for LH/PLOD collagen glycosyltransferase activity
International Journal of Molecular Biosciences, 24, 11213. (2023) *Co-first authors, #Co-second authors - PubMed
A Fe2+-dependent self-inhibited state influences the druggability of human collagen lysyl hydroxylase (LH/PLOD) enzymes
Frontiers in Molecular Biosciences, 9, 876352. (2022) *Co-first authors, **Shared Corresponding Authors
- PubMed
New Mechanistic Insights to PLOD1-mediated Human Vascular Disease
Translational Research, S1931-5244, 00192-00194. (2021)
- PubMed
Full-Length Human Collagen Lysyl Hydroxylases
Encyclopedia of Inorganic and Bioinorganic Chemistry, 2739. (2020) *Equal Contribution
Pathogenic variants in PLOD3 result in a Stickler syndrome-like connective tissue disorder with vascular complications.
Journal of Medical Genetics, 56, 629-638. (2019)
- PubMed
SiMPLOD, a structure-integrated database of collagen lysyl hydroxylase (LH/PLOD) enzyme variants.
Journal of Bone and Mineral Research, 34, 1376-1382. (2019) *Equal Contribution
- PubMed
Molecular architecture of the multifunctional collagen lysyl hydroxylase and glycosyltransferase LH3.
Nature Communications, 9, 3163. (2018)
- PubMed
Autosomal recessive Keratoderma-Ichthyosis-Deafness (ARKID) syndrome is caused by VPS33B mutations affecting Rab protein interaction and collagen modification.
Journal of Investigative Dermatology, S0022-202X, 32800-32807. (2016)
- PubMed
Regulation of post-Golgi LH3 trafficking is essential for collagen homeostasis.
Nature Communications, 7, 12111. (2016) *Shared Corresponding Authors
- PubMed
Press releases (in Italian):
Highlights from the media: Newspapers Articles (in Italian, see more in the Outreach Section):
Active Grants related to this research:
Completed Grants related to this research:
![]() MSCA-IF COTETHERS project, funded by the EU H2020 program
MSCA-IF COTETHERS project, funded by the EU H2020 program
