List of Publications
Nanodecoy systems based on analogues of viral cellular receptors assembled onto fluid lipid-based membranes of nano/extravescicles are potential new tools to complement classic therapeutic or preventive antiviral approaches. The need for lipid-based membranes for transmembrane receptor anchorage may pose technical challenges along industrial translation, calling for alternative geometries for receptor multimerization. Here we developed a semisynthetic self-assembling SARS-CoV-2 nanodecoy by multimerizing the biotin labelled virus cell receptor -ACE2-ectodomain onto a poly-avidin nanoparticle (NP) based on the Avidin-Nucleic-Acid-NanoASsembly-ANANAS. The ability of the assembly to prevent SARS-CoV-2 infection in human lung cells and the affinity of the ACE2:viral receptor-binding domain (RBD) interaction were measured at different ACE2:NP ratios. At ACE2:NP = 30, 90% SARS-CoV-2 infection inhibition at ACE2 nanomolar concentration was registered on both Wuhan and Omicron variants, with ten-fold higher potency than the monomeric protein. Lower and higher ACE2 densities were less efficient suggesting that functional recognition between multi-ligand NPs and multi-receptor virus surfaces requires optimal geometrical relationships. In vivo studies in mice showed that the biodistribution and safety profiles of the nanodecoy are potentially suitable for preventing viral infection upon nasal instillation. Viral receptor multimerization using ANANAS is a convenient process which, in principle, could be rapidly adapted to counteract also other viral infections. Background: Mosquito bite is normally associated with mild allergic responses, but severe localized or systemic reactions are also possible. Reliable tools for the diagnosis of mosquito allergy are still unavailable. Here, we investigated the IgE response to 3 potential salivary allergens identified in the saliva of the tiger mosquito Aedes albopictus. Precise synapse formation is essential for normal functioning of the nervous system. Retinal photoreceptors establish selective contacts with bipolar cells, aligning the neurotransmitter release apparatus with postsynaptic signaling cascades. This involves transsynaptic assembly between the dystroglycan-dystrophin complex on the photoreceptor and the orphan receptor GPR179 on the bipolar cell, which is mediated by the extracellular matrix protein pikachurin (also known as EGFLAM). This complex plays a critical role in the synaptic organization of photoreceptors and signal transmission, and mutations affecting its components cause blinding disorders in humans. Here, we investigated the structural organization and molecular mechanisms by which pikachurin orchestrates transsynaptic assembly and solved structures of the human pikachurin domains by x-ray crystallography and of the GPR179-pikachurin complex by single-particle, cryo-electron microscopy. The structures reveal molecular recognition principles of pikachurin by the Cache domains of GPR179 and show how the interaction is involved in the transsynaptic alignment of the signaling machinery. Together, these data provide a structural basis for understanding the synaptic organization of photoreceptors and ocular pathology. Hydroxylysine glycosylations are post-translational modifications (PTMs) essential for the maturation and homeostasis of fibrillar and non-fibrillar collagen molecules. The multifunctional collagen lysyl hydroxylase 3 (LH3/PLOD3) and the collagen galactosyltransferase GLT25D1 are the human enzymes identified as responsible for the glycosylation of collagen lysines, although a precise description of the contribution of each enzyme to these essential PTMs has not been clarified. LH3/PLOD3 is thought to be capable of performing two chemically-distinct collagen glycosyltransferase reactions using the same catalytic site: an inverting beta-1,O-galactosylation of hydroxylysines (Gal-T) and a retaining alpha-1,2-glucosylation of galactosyl hydroxylysines (Glc-T). In this work, we have combined indirect luminescence-based assays with direct mass spectrometry-based assays and molecular structure studies to demonstrate that LH3/PLOD3 only has Glc-T activity and that GLT25D1 only has Gal-T activity. Structure-guided mutagenesis confirmed that the Glc-T activity is defined by key residues in the first-shell environment of the glycosyltransferase catalytic site as well as by long-range contributions from residues within the same glycosyltransferase (GT) domain. By solving the molecular structures and characterizing the interactions and solving the molecular structures of human LH3/PLOD3 in complex with different UDP-sugar analogs, we show how these studies could provide insights for LH3/PLOD3 glycosyltransferase inhibitor development. Collectively, our data provide new tools for the direct investigation of collagen hydroxylysine PTMs and a comprehensive overview of the complex network of shapes, charges and interactions that enable LH3/PLOD3 glycosyltransferase activities, expanding the molecular framework for improved understanding and manipulation of glycosyltransferase functions in biomedical applications. (Between writing and publication of this chapter, the field of computational structural biology has witnessed unprecedented progress due to the implementation of AI-based algorithms for protein structure prediction and computation of macromolecular assemblies. Presently, the approaches proposed in this chapter still provide a solid approach for modeling of protein complexes, nevertheless the authors suggest to combine what described here with the new technologies to achieve the most reliable results.) The recent advances in structural biology, combined with continuously increasing computational capabilities and development of advanced softwares, have drastically simplified the workflow for protein homology modeling. Modeling of individual proteins is nowadays quick and straightforward for a large variety of protein targets, thanks to guided pipelines relying on advanced computational tools and user-friendly interfaces, which have extended and promoted the use of modeling also to scientists not focusing on molecular structures of proteins. Nevertheless, construction of models of multi-protein complexes remains quite challenging for the non-experts, often due to the usage of specific procedures depending on the system under investigation and the need for experimental validation approaches to strengthen the generated output.In this chapter, we provide a brief overview of the approaches enabling generation of multi-protein complex models starting from homology models of individual protein components. Using real-life examples, we include two examples to guide the reader in the generation of homomeric and heteromeric protein models. Programmed cell death protein 1 (PD-1) is an immunoregulatory target which is recognized by different monoclonal antibodies, approved for the therapy of multiple types of cancer. Different anti-PD-1 antibodies display different therapeutic properties and there is a pharmaceutical interest to generate and characterize novel anti-PD-1 antibodies. We screened multiple human antibody phage display libraries to target novel epitopes on the PD-1 surface and we discovered a unique and previously undescribed binding specificity (termed D12) from a new antibody library (termed AMG). The library featured antibody fragments in single-chain fragment variable (scFv) format, based on the IGHV3-23*03 (VH) and IGKV1-39*01 (V?) genes. The D12 antibody was characterized by surface plasmon resonance (SPR), cross-reacted with the Cynomolgus monkey antigen and bound to primary human T cells, as shown by flow cytometry. The antibody blocked the PD-1/PD-L1 interaction in vitro with an EC50 value which was comparable to the one of nivolumab, a clinically approved antibody. The fine details of the interaction between D12 and PD-1 were elucidated by x-ray crystallography of the complex at a 3.5 Å resolution, revealing an unprecedented conformational change at the N-terminus of PD-1 following D12 binding, as well as partial overlap with the binding site for the cognate PD-L1 and PD-L2 ligands which prevents their binding. The results of the study suggest that the expansion of antibody library repertoires may facilitate the discovery of novel binding specificities with unique properties that hold promises for the modulation of PD-1 activity in vitro and in vivo. Neurotrypsin (NT) is a highly specific nervous system multi-domain serine protease best known for its selective processing of the potent synaptic organizer agrin. Its enzymatic activity is thought to influence processes of synaptic plasticity, with its deregulation causing accelerated neuromuscular junction (NMJ) degeneration or contributing to forms of mental retardation. These biological effects are likely to stem from NT-based regulation of agrin signaling. However, dissecting the exact biological implications of NT-agrin interplay is difficult, due to the scarce molecular detail regarding NT activity and NT-agrin interactions. We developed a strategy to reliably produce and purify a catalytically competent engineered variant of NT called NT-mini and a library of C-terminal agrin fragments, with which we performed a thorough biochemical and biophysical characterization of NT enzyme functionality. We studied the regulatory effects of calcium ions and heparin, identified NT's heparin-binding domain, and discovered how zinc ions induce modulation of enzymatic activity. Additionally, we investigated myotube differentiation and hippocampal neuron excitability, evidencing a dose-dependent increase in neuronal activity alongside a negative impact on myoblast fusion when using the active NT enzyme. Collectively, our results provide in vitro and cellular foundations to unravel the molecular underpinnings and biological significance of NT-agrin interactions. Multifunctional human collagen lysyl hydroxylase (LH/PLOD) enzymes catalyze post-translational hydroxylation and subsequent glycosylation of collagens, enabling their maturation and supramolecular organization in the extracellular matrix (ECM). Overexpression of LH/PLODs in the tumor microenvironment results in abnormal accumulation of collagen post-translational modifications which has been recently correlated with increased metastatic progression of a wide variety of solid tumors, making LH/PLODs excellent candidates for prospective treatment of aggressive cancers. The recent years have witnessed significant research efforts to facilitate drug discovery on LH/PLODs, including molecular structure characterizations and development of reliable high-throughput enzymatic assays. Using a combination of biochemistry and in silico studies, we characterized the dual role of Fe2+ as simultaneous cofactor and inhibitor of lysyl hydroxylase activity and studied the effect of a promiscuous Fe2+ chelating agent, 2,2’-bipyridil, broadly considered a lysyl hydroxylase inhibitor. We found that at low concentrations, 2,2’-bipyridil unexpectedly enhances the LH enzymatic activity by reducing the inhibitory effect of excess Fe2+. Together, our results show a fine balance between Fe2+-dependent enzymatic activity and Fe2+-induced self-inhibited states, highlighting exquisite differences between LH/PLODs and related Fe2+, 2-oxoglutarate dioxygenases and suggest that conventional structure-based approaches may not be suited for successful inhibitor development. These insights address outstanding questions regarding druggability of LH/PLOD lysyl hydroxylase catalytic site and provide a solid ground for upcoming drug discovery and screening campaigns. The mosquito proboscis is an efficient microelectromechanical system, which allows the insect to feed on vertebrate blood quickly and painlessly. Its efficiency is further enhanced by the insect saliva, although through unclear mechanisms. Here we describe the initial trigger of an unprecedented feedback signalling pathway in Aedes mosquitoes affecting feeding behaviour. We identified LIPS proteins in the saliva of Aedes mosquitoes that promote feeding in the vertebrate skin. LIPS show a new all-helical protein fold constituted by two domains. The N-terminal domain interacts with a cuticular protein (Cp19) located at the tip of the mosquito labrum. Upon interaction, the morphology of the labral cuticle changes and this modification is most likely sensed by proprioceptive neurons. Our study identifies an additional role of mosquito saliva and underlines that the external cuticle is a possible site of key molecular interactions affecting the insect biology and its vector competence. Recombinant proteins are essential milestones for a plethora of different applications, ranging from pharmaceutical to clinical fields, and mammalian cell lines are amongst the currently preferred systems to obtain large amounts of the proteins of interest, due to their high level of posttranslational modifications as well as manageable large-scale production. In this regard, human embryonic kidney 293 (HEK293) cells constitute one of the main standard lab-scale mammalian host for recombinant protein production, as these cells are relatively easy to handle, scale-up and transfect. Here we present a detailed protocol for the cost-effective, reproducible and scalable implementation of HEK293 cell cultures in suspension, suitable for commercially available HEK293 cells (HEK293-F), for high-quantity recombinant production of secreted soluble multi-domain protein targets. In addition, the protocol is optimized for a Monday-to-Friday maintenance schedule, thus simplifying and streamlining the work of the operators responsible for cell culture maintenance. Collagen is a major constituent of the extracellular matrix (ECM) that confers fundamental mechanical properties to tissues. To allow proper folding in triple-helices and organization in quaternary super-structures, collagen molecules require essential post-translational modifications (PTMs), including hydroxylation of proline and lysine residues, and subsequent attachment of glycan moieties (galactose and glucose) to specific hydroxylysine residues on procollagen alpha chains. The resulting galactosyl-hydroxylysine (Gal-Hyl) and less abundant glucosyl-galactosyl-hydroxylysine (Glc-Gal-Hyl) are amongst the simplest glycosylation patterns found in nature and are essential for collagen and ECM homeostasis. These collagen PTMs depend on the activity of specialized glycosyltransferase enzymes. Although their biochemical reactions have been widely studied, several key biological questions about the possible functions of these essential PTMs are still missing. In addition, the lack of three-dimensional structures of collagen glycosyltransferase enzymes hinders our understanding of the catalytic mechanisms producing this modification, as well as the impact of genetic mutations causing severe connective tissue pathologies. In this mini-review, we summarize the current knowledge on the biochemical features of the enzymes involved in the production of collagen glycosylations and the current state-of-the-art methods for the identification and characterization of this important PTM. The use of immunomodulatory agents for the treatment of cancer is gaining a growing biopharmaceutical interest. Antibody-cytokine fusion proteins, namely immunocytokines, represent a promising solution for the regulation of the immune system at the site of disease. The three-dimensional arrangement of these molecules can profoundly influence their biological activity and pharmacokinetic properties. Structural techniques might provide important insight in the 3D arrangement of immunocytokines. Here, we performed structure investigations on clinical grade fusion proteins L19-IL2, IL12-L19L19 and L19L19-IL2 to elucidate their quaternary organization. Crystallographic characterization of the common L19 antibody fragment at a resolution of 2.0-Å was combined with low-resolution studies of the full-length chimeric molecules using small-angle synchrotron X-ray scattering (SAXS) and negative stain electron microscopy. Characterization of the full-length quaternary structures of the immunocytokines in solution by SAXS consistently supported the diabody structure in the L19-IL2 immunocytokine and allowed generation of low-resolution models of the chimeric proteins L19L19-IL2 and IL12-L19L19. Comparison with 3D reconstructions obtained from negative-stain electron microscopy revealed marked flexibility associated to the linker regions connecting the cytokine and the antibody components of the chimeric proteins. Collectively, our results indicate that low-resolution molecular structure characterizations provide useful complementary insights for the quality control of immunocytokines, constituting a powerful tool to guide the design and the subsequent optimization steps towards clinical enhancement of these chimeric protein reagents. An artwork describing this paper was selected as Front Cover image for the March 2021 issue of the Journal of Structural Biology: Synapse formation is a very elaborate process dependent upon accurate coordination of pre and postsynaptic specialization, requiring multiple steps and a variety of receptors and signaling molecules. Due to its relative structural simplicity and the ease in manipulation and observation, the neuromuscular synapse or neuromuscular junction (NMJ) - the connection between motor neurons and skeletal muscle - represents the archetype junction system for studying synapse formation and conservation. This junction is essential for survival, as it controls our ability to move and breath. NMJ formation requires coordinated interactions between motor neurons and muscle fibers, which ultimately result in the formation of a highly specialized postsynaptic architecture and a highly differentiated nerve terminal. Furthermore, to ensure a fast and reliable synaptic transmission following neurotransmitter release, ligand-gated channels (acetylcholine receptors, AChRs) are clustered on the postsynaptic muscle cell at high concentrations in sites opposite the presynaptic active zone, supporting a direct role for nerves in the organization of the postsynaptic membrane architecture. This organized clustering process, essential for NMJ formation and for life, relies on key signaling molecules and receptors and is regulated by soluble extracellular molecules localized within the synaptic cleft. Notably, several mutations as well as auto-antibodies against components of these signaling complexes have been related to neuromuscular disorders. The recent years have witnessed strong progress in the understanding of molecular identities, architectures and functions of NMJ macromolecules. Among these, prominent roles have been proposed for neural variants of the proteoglycan agrin, its receptor at NMJs composed of the lipoprotein receptor-related protein 4 (LRP4) and the muscle-specific kinase (MuSK), as well as the regulatory soluble synapse-specific protease Neurotrypsin. In this review we summarize the current state of the art regarding molecular structures and (agrin-dependent) canonical, as well as (agrin-independent) non canonical, MuSK signaling mechanisms that underscore the formation of neuromuscular junctions, with the aim of providing a broad perspective to further stimulate molecular, cellular and tissue biology investigations on this fundamental intercellular contact. Current knowledge of the piRNA pathway is based mainly on studies on Drosophila melanogaster where three proteins of the Piwi subclade of the Argonaute family interact with PIWI-interacting RNAs to silence transposable elements in gonadal tissues. In mosquito species that transmit epidemic arboviruses such as dengue and chikungunya viruses, Piwi clade genes underwent expansion, are also expressed in the soma and cross-talk with proteins of recognized antiviral function cannot be excluded for some Piwi proteins. These observations underscore the importance of expanding our knowledge of the piRNA pathway beyond the model organism D. melanogaster. Here we focus on the emerging arboviral vector Aedes albopictus and we couple traditional approaches of expression and adaptive evolution analyses with most current computational predictions of protein structure to study evolutionary divergence among Piwi clade proteins. Superposition of protein homology models indicate possible high structure similarity among all Piwi proteins, with high levels of amino acid conservation in the inner regions devoted to RNA binding. On the contrary, solvent-exposed surfaces showed low conservation, with several sites under positive selection. Analysis of the expression profiles of Piwi transcripts during mosquito development and following infection with Dengue serotype 1 or Chikungunya viruses showed a concerted elicitation of all Piwi transcripts during viral dissemination of dengue viruses while maintenance of infection relied on expression of primarily Piwi5. Opposite, establishment of persistent infection by Chikungunya virus is accompanied by increased expression of all Piwi genes, particularly Piwi4 and, again, Piwi5. Overall these results are consistent with functional specialization and a general antiviral role for Piwi5. Experimental evidences of sites under positive selection in Piwi1-3, Piwi4 and Piwi6, that have complex expression profiles, provide useful knowledge to design tailored functional experiments. BACKGROUND- Taking advantage of several factors, as worldwide trading, climatic changes and urbanization, Aedes mosquitoes are impressively expanding their geographic distribution. A paradigm is provided by the rapid global spreading of Aedes albopictus, a species that is a competent vector of several arboviral diseases (e.g. dengue, Zika, chikungunya) and has been responsible of quite a few outbreaks in the last decade. Historically, vector control always played a pivotal role for the containment of arthropod-borne diseases, and this appears especially crucial for arboviral diseases for which no effective vaccines or specific medications are available. Currently, host exposure to mosquitoes is indirectly evaluated by entomological methods; however, exploitation of human immune responses to mosquito salivary proteins is emerging as a relevant additional tool, with important epidemiological implications for the evaluation of mosquito-borne disease risk. This study provides preliminary but solid indications that the 34k2 salivary proteins from Ae. albopictus and Aedes aegypti may be suitable candidates for the development of serological assays to evaluate spatial and/or temporal variation of human exposure to Aedes vectors. Combined to the presently available tools to assess arboviral exposure/infection, this may be of great help for the development of a serological toolbox allowing for the simultaneous determination of human exposure to Aedes vectors and to the pathogens they transmit. BACKGROUND- Pathogenic PLOD3 variants cause a connective tissue disorder (CTD) that has been described rarely. We further characterise this CTD and propose a clinical diagnostic label to improve recognition and diagnosis of PLOD3-related disease. An artwork describing this paper was selected for the cover of this issue of the Journal of Medical Genetics: LSD1 and LSD2 are homologous histone demethylases with opposite biological outcomes related to chromatin silencing and transcription elongation, respectively. Unlike LSD1, LSD2 nucleosome-demethylase activity relies on a specific linker peptide from the multidomain protein NPAC. We used single-particle cryoelectron microscopy (cryo-EM), in combination with kinetic and mutational analysis, to analyze the mechanisms underlying the function of the human LSD2/NPAC-linker/nucleosome complex. Weak interactions between LSD2 and DNA enable multiple binding modes for the association of the demethylase to the nucleosome. The demethylase thereby captures mono- and dimethyl Lys4 of the H3 tail to afford histone demethylation. Our studies also establish that the dehydrogenase domain of NPAC serves as a catalytically inert oligomerization module. While LSD1/CoREST forms a nucleosome docking platform at silenced gene promoters, LSD2/NPAC is a multifunctional enzyme complex with flexible linkers, tailored for rapid chromatin modification, in conjunction with the advance of the RNA polymerase on actively transcribed genes. Neurotrypsin (NT) is a multi-domain serine protease of the nervous system with only one known substrate: the large proteoglycan Agrin. NT has seen to be involved in the maintenance/turnover of neuromuscular junctions and in processes of synaptic plasticity in the central nervous system. Roles which have been tied to its enzymatic activity, localized in the C-terminal serine-protease (SP) domain. However the purpose of NT's remaining 3-4 scavenger receptor cysteine-rich (SRCR) domains is still unclear. We have determined the crystal structure of the third SRCR domain of murine NT (mmNT-SRCR3), immediately preceding the SP domain and performed a comparative structural analysis using homologous SRCR structures. Our data and the elevated degree of structural conservation with homologous domains highlight possible functional roles for NT SRCRs. Computational and experimental analyses suggest the identification of a putative binding region for Ca2+ ions, known to regulate NT enzymatic activity. Furthermore, sequence and structure comparisons allow to single out regions of interest that, in future studies, might be implicated in Agrin recognition/binding or in interactions with as of yet undiscovered NT partners. PLOD genes encode for procollagen lysyl hydroxylase enzymes (LH/PLOD), a family of proteins essential for collagen biosynthesis. Several mutations affect these genes causing severe disorders, such as Ehlers-Danlos and Bruck syndrome, as well a connective tissue disease with phenotype resembling osteogenesis imperfecta caused by lack of LH3/PLOD3 functions. The recently determined three-dimensional structures of the full-length human LH3/PLOD3 isoform, together with the structure of a fragment of a viral LH/PLOD homolog, are now allowing molecular mapping of the numerous disease-causing mutations, providing insights often suitable for the interpretation of the resulting disease phenotypes. However, the added value of molecular structure interpretation is affected by the limited accessibility of complex molecular data to scientific communities lacking direct expertise in structural biology. In this work, we present SiMPLOD (Structurally-integrated database for Mutations of PLOD genes), a publicly-available manually-curated online database with an embedded molecular viewer interface for the visualization and interpretation of LH/PLOD mutations on available molecular models. Each SiMPLOD entry is accompanied by manual annotations extrapolated from literature references and comments about the localization of the amino acid variants on the molecular structure. Additional links to the appropriate online resources for clinically-relevant as well as biochemical data are also provided in a standardized format. The web application is available at http://fornerislab.unipv.it/SiMPLOD. In insects, odorant?binding proteins (OBPs) connect the peripheral sensory system to receptors of olfactory organs. Medfly Ceratitis capitata CcapObp22 shows 37% identity and close phylogenetic affinities with Drosophila melanogaster OBP69a/PB?PRP1 (pheromone?binding protein related protein 1). The CcapObp22 gene is transcribed in the antennae and maxillary palps, suggesting an active role in olfaction. Here, we recombinantly produced CcapObp22, obtaining a 13.5 kDa protein capable of binding multiple strongly hydrophobic terpene compounds, including medfly male pheromone components. The highest binding affinity (EC50 0.48 ?M) was to (E,E)???farnesene, one of the most abundant compounds in the male pheromone blend. This odorant was used in co?crystallization experiments, yielding the structure of CcapOBP22. The monomeric structure shows the typical OBP folding, constituted by six ??helical elements interconnected by three disulfide bridges. A C?terminal seventh ??helix constitutes the wall of a deep, L?shaped hydrophobic cavity. Analysis of the electron density in this cavity suggested trapping of farnesene in the crystal structure, although with partial occupancy. Superposition of the CcapOBP22 structure with related 7?helical OBPs highlights striking similarity in the organization of the C?terminal segment of these proteins. Collectively, our molecular and physiological data on medfly CcapOBP22 suggest its involvement in inter?sex olfactory communication. Lysyl hydroxylases catalyze hydroxylation of collagen lysines, and sustain essential roles in extracellular matrix (ECM) maturation and remodeling. Malfunctions in these enzymes cause severe connective tissue disorders. Human lysyl hydroxylase 3 (LH3/PLOD3) bears multiple enzymatic activities, as it catalyzes collagen lysine hydroxylation and also their subsequent glycosylation. Our understanding of LH3 functions is currently hampered by lack of molecular structure information. Here, we present high resolution crystal structures of full-length human LH3 in complex with cofactors and donor substrates. The elongated homodimeric LH3 architecture shows two distinct catalytic sites at the N- and C-terminal boundaries of each monomer, separated by an accessory domain. The glycosyltransferase domain displays distinguishing features compared to other known glycosyltransferases. Known disease-related mutations map in close proximity to the catalytic sites. Collectively, our results provide a structural framework characterizing the multiple functions of LH3, and the molecular mechanisms of collagen-related diseases involving human lysyl hydroxylases. Press Release (in Italian) - Newspapers Articles (in Italian): 1, 2, 3, 4, 5 Quorum sensing (QS) is a bacterial intercellular communication process which controls the production of major virulence factors, such as proteases, siderophores, and toxins, as well as biofilm formation. Since the inhibition of this pathway reduces bacterial virulence, QS is considered a valuable candidate drug target, particularly for the treatment of opportunistic infections, such as those caused by Burkholderia cenocepacia in cystic fibrosis patients. Diketopiperazine inhibitors of the acyl homoserine lactone synthase CepI have been recently described. These compounds are able to impair the ability of B. cenocepacia to produce proteases, siderophores, and to form biofilm, being also active in a Caenorhabditis elegans infection model. However, the precise mechanism of action of the compounds, as well as their effect on the cell metabolism, fundamental for candidate drug optimization, are still not completely defined. Here, we performed a proteomic analysis of B. cenocepacia cells treated with one of these inhibitors, and compared it with a cepI deleted strain. Our results demonstrate that the effects of the compound are similar to the deletion of cepI, clearly confirming that these molecules function as inhibitors of the acyl homoserine lactone synthase. Moreover, to deepen our knowledge about the binding mechanisms of the compound to CepI, we exploited previously published in silico structural insights about this enzyme structure and validated different candidate binding pockets on the enzyme surface using site-directed mutagenesis and biochemical analyses. Our experiments identified a region near the predicted S-adenosylmethionine binding site critically involved in interactions with the inhibitor. These results could be useful for future structure-based optimization of these CepI inhibitors. Burkholderia cenocepacia, an opportunistic respiratory pathogen particularly relevant for cystic fibrosis patients, is difficult to eradicate due to its high level of resistance to most clinically relevant antimicrobials. Consequently, the discovery of new antimicrobials as well as molecules capable of inhibiting its virulence is mandatory. In this regard quorum sensing (QS) represents a good target for anti-virulence therapies, as it has been linked to biofilm formation and is important for the production of several virulence factors, including proteases and siderophores. Here, we report the discovery of new diketopiperazine inhibitors of the B. cenocepacia acyl homoserine lactone synthase CepI, and report their anti-virulence properties. Out of ten different compounds assayed against recombinant CepI, four were effective inhibitors, with IC50 values in the micromolar range. The best compounds interfered with protease and siderophore production, as well as with biofilm formation, and showed good in vivo activity in a Caenorhabditis elegans infection model. These molecules were also tested in human cells and showed very low toxicity. Therefore, they could be considered for in vivo combined treatments with established or novel antimicrobials, to improve the current therapeutic strategies against B. cenocepacia. Structural biology comprises a variety of tools to obtain atomic resolution data for the investigation of macromolecules. Conventional structural methodologies including crystallography, NMR and electron microscopy often do not provide sufficient details concerning flexibility and dynamics, even though these aspects are critical for the physiological functions of the systems under investigation. However, the increasing complexity of the molecules studied by structural biology (including large macromolecular assemblies, integral membrane proteins, intrinsically disordered systems, and folding intermediates) continuously demands in-depth analyses of the roles of flexibility and conformational specificity involved in interactions with ligands and inhibitors. The intrinsic difficulties in capturing often subtle but critical molecular motions in biological systems have restrained the investigation of flexible molecules into a small niche of structural biology. Introduction of massive technological developments over the recent years, which include time-resolved studies, solution X-ray scattering, and new detectors for cryo-electron microscopy, have pushed the limits of structural investigation of flexible systems far beyond traditional approaches of NMR analysis. By integrating these modern methods with powerful biophysical and computational approaches such as generation of ensembles of molecular models and selective particle picking in electron microscopy, more feasible investigations of dynamic systems are now possible. Using some prominent examples from recent literature, we review how current structural biology methods can contribute useful data to accurately visualize flexibility in macromolecular structures and understand its important roles in regulation of biological processes. Post-translational modifications are necessary for collagen precursor molecules (procollagens) to acquire final shape and function. However, the mechanism and contribution of collagen modifications that occur outside endoplasmic reticulum and Golgi are not understood. We discovered that VIPAR, with its partner proteins, regulate sorting of Lysyl Hydroxylase 3 (LH3, also known as PLOD3) into newly-identified post-Golgi collagen IV carriers and that VIPAR-dependent sorting is essential for modification of lysines in multiple collagen types. Identification of structural and functional collagen abnormalities in cells and tissues from patients and murine models of the autosomal recessive multisystem disorder Arthrogryposis, Renal dysfunction and Cholestasis syndrome caused by VIPAR and VPS33B deficiencies confirmed our findings. Thus, regulation of post-Golgi LH3 trafficking is essential for collagen homeostasis and for the development and function of multiple organs and tissues. Press Release (in Italian) - Newspapers Articles (in Italian): 1, 2 Burkholderia cenocepacia is a major concern among respiratory tract infections in cystic fibrosis patients. This pathogen is particularly difficult to treat because of its high level of resistance to the clinically relevant antimicrobial agents. In B. cenocepacia, the quorum sensing cell-cell communication system is involved in different processes important for the bacterial virulence, such as biofilm formation or protease and siderophore production. Targeting the enzymes involved in this process represents a promising therapeutic approach. With the aim of finding effective quorum sensing inhibitors, we have determined the three-dimensional structure of B. cenocepacia diffusible factor synthase A, DfsA. This bifunctional crotonase (dehydratase/thioesterase) produces the characteristic quorum sensing molecule of B. cenocepacia, cis-2-dodecenoic acid or BDSF, starting from 3-hydroxydodecanoyl-acyl carrier protein. Unexpectedly, the crystal structure revealed the presence of a lipid molecule into the catalytic site of the enzyme, which was identified as dodecanoic acid. Our biochemical characterization shows that DfsA is able to use dodecanoyl-acyl carrier protein as a substrate, demonstrating that dodecanoic acid, the product of this reaction, is released very slowly from the DfsA active site, therefore acting as a DfsA inhibitor. This molecule shows an unprecedented conformational arrangement inside the DfsA active site. In contrast with previous hypotheses, our data illustrate how DfsA and closely-related homologous enzymes can recognize long hydrophobic substrates without large conformational changes or assistance by additional regulator molecules. The elucidation of the substrate binding mode in DfsA provides the starting point for structure-based drug discovery studies targeting B. cenocepacia quorum sensing-assisted virulence. Regulators of complement activation (RCA) inhibit complement-induced immune responses on healthy host tissues. We present crystal structures of humanRCA(MCP, DAF, andCR1) and a smallpox virus homolog (SPICE) bound to complement component C3b. Our structural data reveal that up to four consecutive homologousCCPdomains (i-iv), responsible for inhibition, bind in the same orientation and extended arrangement at a shared binding platform on C3b. Large sequence variations inCCPdomains explain the diverse C3b-binding patterns, with limited or no contribution of some individual domains, while all regulators show extensive contacts with C3b for the domains at the third site. A variation of ~100° rotation around the longitudinal axis is observed for domains binding at the fourth site on C3b, without affecting the overall binding mode. The data suggest a common evolutionary origin for both inhibitory mechanisms, called decay acceleration and cofactor activity, with variable C3b binding through domains at sites ii, iii, and iv, and provide a framework for understanding RCA disease-related mutations and immune evasion. Hydroxysteroid dehydrogenases are of great interest as biocatalysts for transformations involving steroid substrates. They feature a high degree of stereo- and regio-selectivity, acting on a defined atom with a specific configuration of the steroid nucleus. The crystal structure of 7?-hydroxysteroid dehydrogenase from Collinsella aerofaciens reveals a loop gating active-site accessibility, the bases of the specificity for NADP+, and the general architecture of the steroid binding site. Comparison with 7?-hydroxysteroid dehydrogenase provides a rationale for the opposite stereoselectivity. The presence of a C-terminal extension reshapes the substrate site of the ?-selective enzyme, possibly leading to an inverted orientation of the bound substrate. An image from our paper was selected as back cover image for this issue of proteins (Page C4): The recently discovered cytokinin (CK)-specific phosphoribohydrolase Lonely Guy (LOG) is a key enzyme of CK biosynthesis, converting inactive CK nucleotides into biologically active free bases. We have determined the crystal structures of LOG from Claviceps purpurea (cpLOG) and its complex with the enzymatic product phosphoribose. The structures reveal a dimeric arrangement of Rossmann folds, with the ligands bound to large pockets at the interface between cpLOG monomers. Structural comparisons highlight the homology of cpLOG to putative lysine decarboxylases. Extended sequence analysis enabled identification of a distinguishing LOG sequence signature. Taken together, our data suggest phosphoribohydrolase activity for several proteins of unknown function.2025

Piccinno, R., Fiorenza, G., Vasquez, M.I., Bouyer, J., Notarides, G., Gomulski, L.M., Meletiou, S., Akiner, M., Michaelakis, A., Forneris, F., Maga, G., Gasperi, G., Malacrida, A.R.
On the tracks of an uninvited guest, the Asian tiger mosquito, Aedes albopictus in Cyprus
Parasites and Vectors, 18, 39. (2025) - PubMed
2024

Brambilla, L., Valori, C.F., Guidotti, G., Martorana, F., Sulmona, C., De Martini, L.B., Canciani, A., Fumagalli, M., Talpo, F., Biella, G., di Pasquale, E., Iacobucci, C., Forneris, F., Zhou, H., Rossi D.
Recombinant SMN protein synergizes with spinal muscular atrophy therapy to counteract pathological motor neuron phenotypes
Translational Neurodegeneration, 13, 63. (2024) - PubMed

Valeri, E., Breggion, S., Barzaghi, F., Abou Alezz, M., Pagani, I., Forneris, F., Sartirana, C., Costantini, M., Costi, S., Marino, A., Chiarotto, E., Colavito, D., Cimaz, R., Merelli, I., Vicenzi, E., Aiuti, A., Kajaste-Rudnitski, A.
A novel STING variant triggers endothelial toxicity and SAVI disease
Journal of Experimental Medicine, 221, e20232167. (2024) - PubMed

Perico, L., Casiraghi, F., Sônego, F., Todeschini, M., Corna, D., Cerullo, D., Pezzotta, A., Isnard-Petit, P., Faravelli, S., Forneris, F., Thiam, K., Benigni, A., Remuzzi, G.
Bi-specific Autoantigen-T cell Engagers as targeted immunotherapy for autoreactive B cell depletion in autoimmune diseases
Frontiers in Immunology, 15, 1335998. (2024) - PubMed
2023

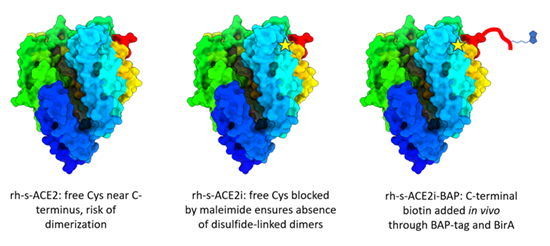
Bernardotto S., Frasson I., Faravelli S., Morelli A., Schiavon E., Moscatiello G.Y., Violatto M.B., Pinnola A., Canciani A., Mattarei A., Rossi G., Brini M., Pasetto L., Bonetto V., Bigini P., Forneris F., Richter S.N., Morpurgo M.
Efficient SARS-CoV-2 infection antagonization by rhACE2 ectodomain multimerized onto the avidin nucleic-acid-NanoASsembly
Biomaterials, 303, 122394. (2023) - PubMed

Arnoldi I., Villa M., Mancini G., Varotto-Boccazzi I., Yacoub M.R., Asperti C., Mascheri A., Casiraghi S., Epis S., Bandi C., Dagna L., Forneris F., Gabrieli P.
IgE response to Aed al 13 and Aed al 14 recombinant allergens from Aedes albopictus saliva in humans
World Allergy Organization Journal, 16, 100836. (2023) - PubMed
Methods: Serum from 55 adult individuals (28 controls and 27 allergic people), were analysed using an in-house Enzyme Linked ImmunoSorbent Assay (ELISA) against the Salivary Gland Extract (SGE) and the recombinant proteins albD7l2 (Aed al 2), albAntigen5-3 (Aed al 13) and albLIPS-2 (Aed al 14).
Results: Fifteen of the 27 (56%) individuals having hypersensitive reactions to mosquito bites had IgE serum levels recognizing SGE. Negative sera did not show detectable levels of IgE targeting the SGE from the most common sympatric mosquito Culex pipiens. Among the positive individuals, 2 subjects displayed IgE targeting Aed al 2 (13%), while IgE recognizing Aed al 13 and Aed al 14 were detected in ten (67%) and seven (47%) individuals, respectively. Two sera from non-hypersensitive subjects had detectable levels of IgE targeting Aed al 13, suggesting possible cross-reaction with the homologue salivary proteins of multiple mosquito species or, more generally, of hematophagous insects.
Conclusions: Our results indicate that Aed al 13 and Aed al 14 hold the potential to be developed as tools for the diagnosis of allergy to Ae. albopictus bites. Such tools would facilitate epidemiological studies on tiger mosquito allergy in humans and might foster the development of further protein-based assays to investigate cross-species allergies.

Mangiacotti, M.*,#, Fumagalli, M.*, Casali, C., Biggiogera, M., Forneris, F., Sacchi, R.
Carbonic anhydrase IV in lizard chemical signals
Scientific Reports, 13, 14164. (2023) - PubMed
*Equal Contribution - #Corresponding Author
Patil, D.N.*, Pantalone, S., Cao, Y., Laboute, T., Novick, S.J., Singh, S., Savino, S., Faravelli, S., Magnani, F., Griffin, P.R., Singh, A.K., Forneris, F.*, Martemyanov, K.A.*
Structure of the photoreceptor synaptic assembly of the extracellular matrix protein pikachurin with the orphan receptor GPR179
Science Signaling, 16, eadd9539. (2023) - PubMed
*Shared Corresponding Authors 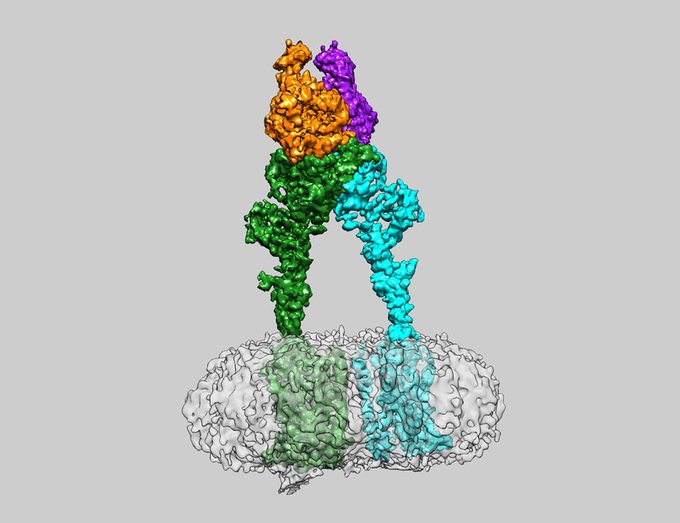
Mattoteia, D.*, Chiapparino, A.*, Fumagalli, M.**, De Marco, M.**, De Giorgi, F.**, Negro, L., Pinnola, A., Faravelli, S., Roscioli, T., Scietti, L., Forneris, F.
Identification of regulatory molecular "hot spots" for LH/PLOD collagen glycosyltransferase activity
International Journal of Molecular Sciences, 24, 11213. (2023) - PubMed
*Co-First authors; **Co-Second authors 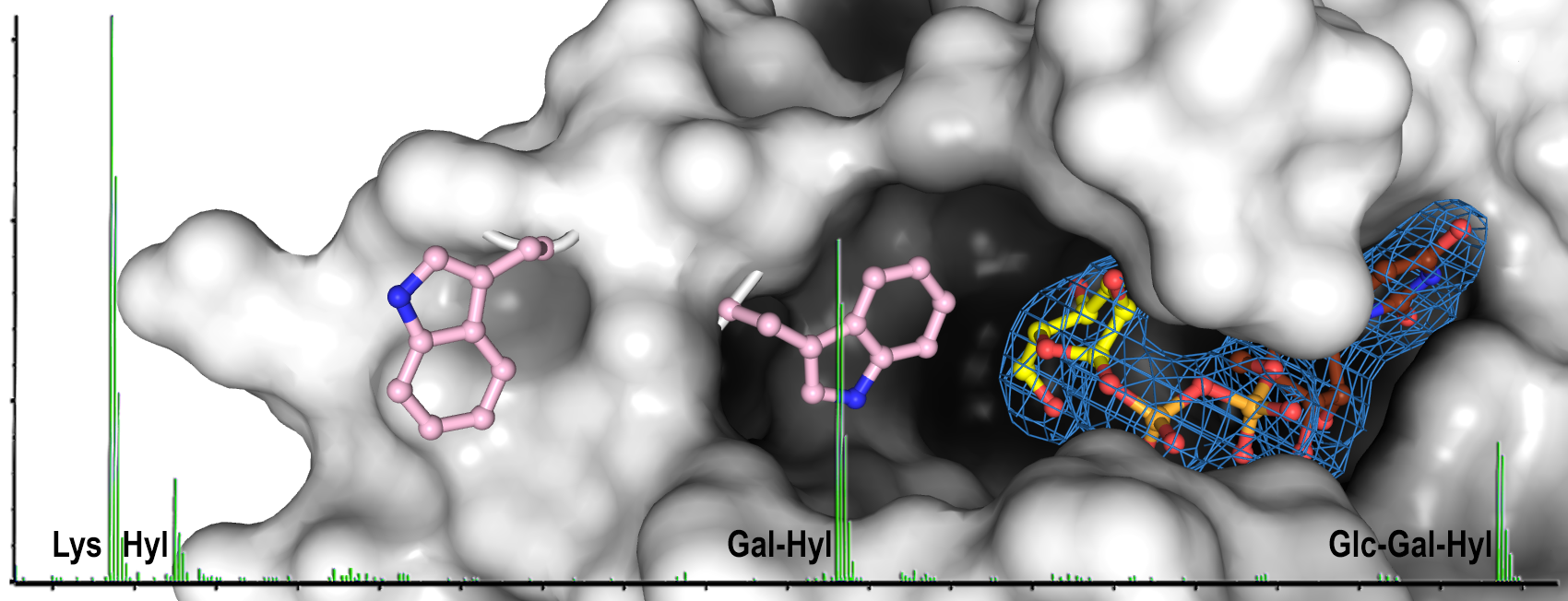

Scietti, L., Forneris, F.
Modeling of Protein Complexes
Methods in Molecular Biology, 2627, 349-371. (2023) - PubMed
2022

Peissert, F.*, Plüss L.*, Giudice A.M.*, Ongaro T., Villa A., Elsayed A., Nadal L., Dakhel Plaza S., Scietti L., Puca E., De Luca R., Forneris F.**, Neri D.**
Selection of a PD-1 blocking antibody from a novel fully human phage display library
Protein Science, 31, e4486. (2022) - PubMed
*Equal Contribution - **Shared Corresponding Authors
Canciani A.*, Capitanio C., Stanga, S., Faravelli S., Scietti L., Mapelli L., Soda T., D'Angelo E., Kienlen-Campard P., Forneris F.*
Deconstruction of Neurotrypsin Reveals a Multi-factorially Regulated Activity Affecting Myotube Formation and Neuronal Excitability
Molecular Neurobiology, 59, 7466-7485. (2022) - PubMed
*Shared Corresponding Authors

Brasu N., Elia I., Russo V., Montacchiesi G., Stabile S.A., De Intinis C., Fesi F., Gizzi K., Macagno M., Montone M., Mussolin B., Grifoni A., Faravelli S., Marchese S., Forneris F., De Francesco R., Sette A., Barnaba V., Sottile A., Sapino A., Pace L.
Memory CD8+ T cell diversity and B cell responses correlate with protection against SARS-CoV-2 following mRNA vaccination
Nature Immunology, 23, 1445-1456. (2022) - PubMed

Scietti L.*,**, Moroni E.*, Mattoteia D.*, Fumagalli M., De Marco M., Negro L., Chiapparino A., Serapian S.A., De Giorgi F., Faravelli S., Colombo G., Forneris F.**
A Fe2+-dependent self-inhibited state influences the druggability of human collagen lysyl hydroxylase (LH/PLOD) enzymes
Frontiers in Molecular Biosciences, 9, 876352. (2022) - PubMed
*Equal Contribution - **Shared Corresponding Authors
De Nola G., Leclerq B., Mougel A., Taront S., Simonneau C., Forneris F., Adriaenssens E., Drobecq H., Iamele L., Dubuquoy L., Melnyk O., Gherardi E., de Jonge H., Vicogne J.
Dimerization of kringle 1 domain from hepatocyte growth factor/scatter factor provides a potent MET receptor agonist
Life Science Alliance, 5, e202201424. (2022) - PubMed

Arnoldi I.*, Mancini G.*, Fumagalli M., Gastaldi D., D'Andrea L., Bandi C., Di Venere M., Iadarola P., Forneris F.**, Gabrieli P.**
A salivary factor binds a cuticular protein and modulates biting by inducing morphological changes in the mosquito labrum
Current Biology, 32, 3493-3504.e11. (2022) - PubMed
*Equal Contribution - **Shared Corresponding Authors - Dispatch From A. Paige and L. Duvall in Current Biology

Guarino S., Di Bello A., Palamini M., Capillo M.C., Forneris F.
Crystal structure of the kringle domain of human receptor tyrosine kinase-like orphan receptor 1 (hROR1)
Acta Crystallographica section F - Structural Biology Communications, F78, 185-192. (2022) - PubMed
2021

Varotto-Boccazzi, I., Manenti, A., Dapporto, F., Gourlay, L.J., Bisaglia, B., Gabrieli, P., Forneris, F., Faravelli, S., Bollati, V., Rubolini, D., Zuccotti, G., Montomoli, E., Epis1, S., Bandi, C.
Epidemic Preparedness: Leishmania tarentolae as an Easy-to-Handle Tool to Produce Antigens for Viral Diagnosis: Application to COVID-19
Frontiers in Microbiology, 12, 736530. (2021) - PubMed

Koenig, S.N., Cavus, O., Williams, J., Bernier, M., Tonniges, J., Sucharki, H., Dew, T., Akel, M., Baker, P., Madiai, F., De Giorgi, F., Scietti, L., Faravelli, S., Forneris, F., Mohler, P.J., Bradley, E.A.
New Mechanistic Insights to PLOD1-mediated Human Vascular Disease
Translational Research, S1931-5244, 00192-00194. (2021) - PubMed

Pradella, D., Deflorian, G., Pezzotta, A., Di Matteo, A., Belloni, E., Campolungo, D., Paradisi, A., Bugatti, M., Vermi, W., Campioni, M., Chiapparino, A., Scietti, L., Forneris, F., Giampietro, C., Volf, N., Rehman, M., Zacchigna, S., Paronetto, M.P., Pistocchi, A., Eichmann, A., Mehlen, P., Ghigna, C.
A ligand-insensitive UNC5B splicing isoform regulates angiogenesis by promoting apoptosis
Nature Communications, 12, 4872. (2021) - PubMed
Brondino, N., Bertoglio, F., Forneris, F., Faravelli, S., Borghesi, A., Damiani, S., Provenzani, U., Nola, M., Olivola, M., Caviglia, M., Politi, P., Fusar-Poli, L., Fusar-Poli, P.
A Pilot Study on Covid and Autism: Prevalence, Clinical Presentation and Vaccine Side Effects
Brain Sciences, 11, 860. (2021) - PubMed
Faravelli, S.*, Campioni, M., Palamini, M., Canciani, A., Chiapparino, A., Forneris, F.*
Optimized Recombinant Production of Secreted Proteins Using Human Embryonic Kidney (HEK293) Cells Grown in Suspension
Bio-Protocol, 11, e3998. (2021) - PubMed
*Shared Corresponding Authors

De Giorgi, F., Fumagalli, M., Scietti, L.*, Forneris, F.*
Collagen hydroxylysine glycosylation: non-conventional substrates for atypical glycosyltransferase enzymes
Biochemical Society Transactions, 49, 855-866. (2021) - PubMed
*Shared Corresponding Authors

Ongaro, T.*, Guarino, S.R.*, Scietti, L., Palamini, M., Wulhfard, S., Neri, D., Villa, A.**, Forneris, F.**
Inference of molecular structure for characterization and improvement of clinical grade immunocytokines
Journal of Structural Biology, 213, 107696. (2021) - PubMed
Cover Image - *Equal Contribution - **Shared Corresponding Authors 
Cover caption:To effectively “invade” the tumor microenvironment, immunocytokines must overcome physicochemical challenges as in the “space invaders” videogame. Optimization of the immunocytokine format is essential for their performance. In this issue of the Journal of Structural Biology, on page 107696, Ongaro, Guarino et al. discuss the inference of structural biology for quality control and rational design of immunocytokine therapeutic agents.
2020
Bruni, M., Cecatiello, V., Diaz-Basabe, A., Lattanzi, G., Mileti, E., Monzani, S., Pirovano, L., Rizzelli, F., Visintin, C., Bonizzi, G., Giani, M., Lavitrano, M.L., Faravelli, S., Forneris, F., Caprioli, F., Pelicci, P.G., Natoli, G., Pasqualato, S., Mapelli, M., Facciotti, F.
Persistence of Anti-SARS-CoV-2 Antibodies in Non-Hospitalized COVID-19 Convalescent Health Care Workers
Journal of Clinical Medicine, 9, 3188. (2020) - PubMed

Buezo Montero S., Gabrieli P., Montarsi F., Borean A., Capelli S., De Silvestro G., Forneris F., Pombi M., Breda A., Capelli G., Arcà B.
IgG Antibody Responses to the Aedes albopictus 34k2 Salivary Protein as Novel Candidate Marker of Human Exposure to the Tiger Mosquito
Frontiers in Cellular and Infection Microbiology, 10, 377. (2020) - PubMed

Selvanathan A., Nixon C.Y., Zhu Y., Scietti L., Forneris F., Moreno Uribe L.M., Lidral A.C. Jezewski P.A., Mulliken J.B., Murray J.C., Buckley M.F., Cox T.C., Roscioli T.
CDH1 Mutation Distribution and Type Suggests Genetic Differences Between the Etiology of Orofacial Clefting and Gastric Cancer
Genes, 11, 391. (2020) - PubMed

Guarino S.*, Canciani A.*, Forneris F.
Dissecting the Extracellular Complexity of Neuromuscular Junctions Organizers
Frontiers in Molecular Biosciences, 6, 156. (2020) - PubMed
*Equal Contribution
2019

Marconcini M., Hernandez L., Iovino G., Houé V., Valerio F., Palatini U., Pischedda E., Crawford J., White B.J., Lin T., Carballar-Lejarazu R., Ometto L., Forneris F., Failloux A-B., Bonizzoni M.
Polymorphism analyses and protein modelling inform on functional specialization of Piwi clade genes in the arboviral vector Aedes albopictus
PLoS Neglected Tropical Diseases, 13, e0007919. (2019) - PubMed

Buezo Montero S., Gabrieli P., Severini F., Picci L., Di Luca M., Forneris F., Facchinelli L., Ponzi M., Lombardo F., Arcà B.
Analysis in a murine model points to IgG responses against the 34k2 salivary proteins from Aedes albopictus and Aedes aegypti as novel promising candidate markers of host exposure to Aedes mosquitoes
PLoS Neglected Tropical Diseases, 13, e0007806. (2019) - PubMed

Ewans L.J., Colley A., Gaston-Massuet C., Gualtieri A., Cowley M.J., McCabe M.J., Anand D., Lachke S., Scietti L., Forneris F., Zhu Y., Ying K., Walsh C., Kirk E.P., Miller D., Giunta C., Sillence D., Dinger M., Buckley M., Roscioli T.
Pathogenic variants in PLOD3 result in a Stickler syndrome-like connective tissue disorder with vascular complications
Journal of Medical Genetics, 56, 629-638. (2019) - PubMed
METHODS- Reported PLOD3 phenotypes were compared with known CTDs utilising data from three further individuals from a consanguineous family with a homozygous PLOD3 c.809C>T; p.(Pro270Leu) variant. PLOD3 mRNA expression in the developing embryo was analysed for tissue-specific localisation. Mouse microarray expression data were assessed for phylogenetic gene expression similarities across CTDs with overlapping clinical features.
RESULTS- Key clinical features included ocular abnormalities with risk for retinal detachment, sensorineural hearing loss, reduced palmar creases, finger contractures, prominent knees, scoliosis, low bone mineral density, recognisable craniofacial dysmorphisms, developmental delay and risk for vascular dissection. Collated clinical features showed most overlap with Stickler syndrome with variable features of Ehlers-Danlos syndrome (EDS) and epidermolysis bullosa (EB). Human lysyl hydroxylase 3/PLOD3 expression was localised to the developing cochlea, eyes, skin, forelimbs, heart and cartilage, mirroring the clinical phenotype of this disorder.
CONCLUSION- These data are consistent with pathogenic variants in PLOD3 resulting in a clinically distinct Stickler-like syndrome with vascular complications and variable features of EDS and EB. Early identification of PLOD3 variants would improve monitoring for comorbidities and may avoid serious adverse ocular and vascular outcomes.
Cover caption:A pathogenic variant in PLOD3 associated with a Stickler syndrome-like connective tissue disorder. See Ewans et al, pages 631, 632.

Marabelli C.*, Marrocco B.*, Pilotto S:*, Chittori S.*, Picaud S., Marchese S., Ciossani G., Forneris F., Filippakopoulos P., Schoehn G., Rhodes D., Subramaniam S.**, Mattevi A.**
A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex
Cell Reports, 27, 387-399. (2019) - PubMed
*Equal Contribution - **Shared Corresponding Authors

Angiolini F.*, Belloni E.*, Giordano M.*, Campioni M., Forneris F., Paronetto M.P., Lupia, M. Brandas C., Pradella D., Di Matteo A., Giampietro C., Jodice G., Luise C., Bertalot G., Freddi S., Malinverno M., Irimia M., Moulton J., Summerton J., Chiapparino A., Ghilardi C., Giavazzi R., Nyqvist D., Gabellini D., Dejana E., Cavallaro U., Ghigna C.
A Novel L1 Isoform with Angiogenic Activity Generated by NOVA2-mediated Alternative Splicing
eLife, 8, e44305. (2019) - PubMed
*Equal Contribution

Canciani A., Catucci G., Forneris F.
Structural characterization of the third scavenger receptor cysteine-rich domain of murine Neurotrypsin
Protein Science, 28, 746-755. (2019) - PubMed

Scietti L.*, Campioni M.*, Forneris F.
SiMPLOD, a structure-integrated database of collagen lysyl hydroxylase (LH/PLOD) enzyme variants
Journal of Bone and Mineral Research, 34, 1376-1382. (2019) - PubMed
*Equal Contribution - Explore SiMPLOD

Falchetto M.*, Ciossani G.*, Scolari F., Di Cosimo A., Nenci S., Field L.M., Mattevi A., Zhou J.-J., Gasperi G.**, Forneris F.**
Structural and biochemical evaluation of Ceratitis capitata OBP22 affinity for odorants involved in inter-sex communication
Insect Molecular Biology, 28, 431-443. (2019) - PubMed
*Equal Contribution - **Shared Corresponding Authors 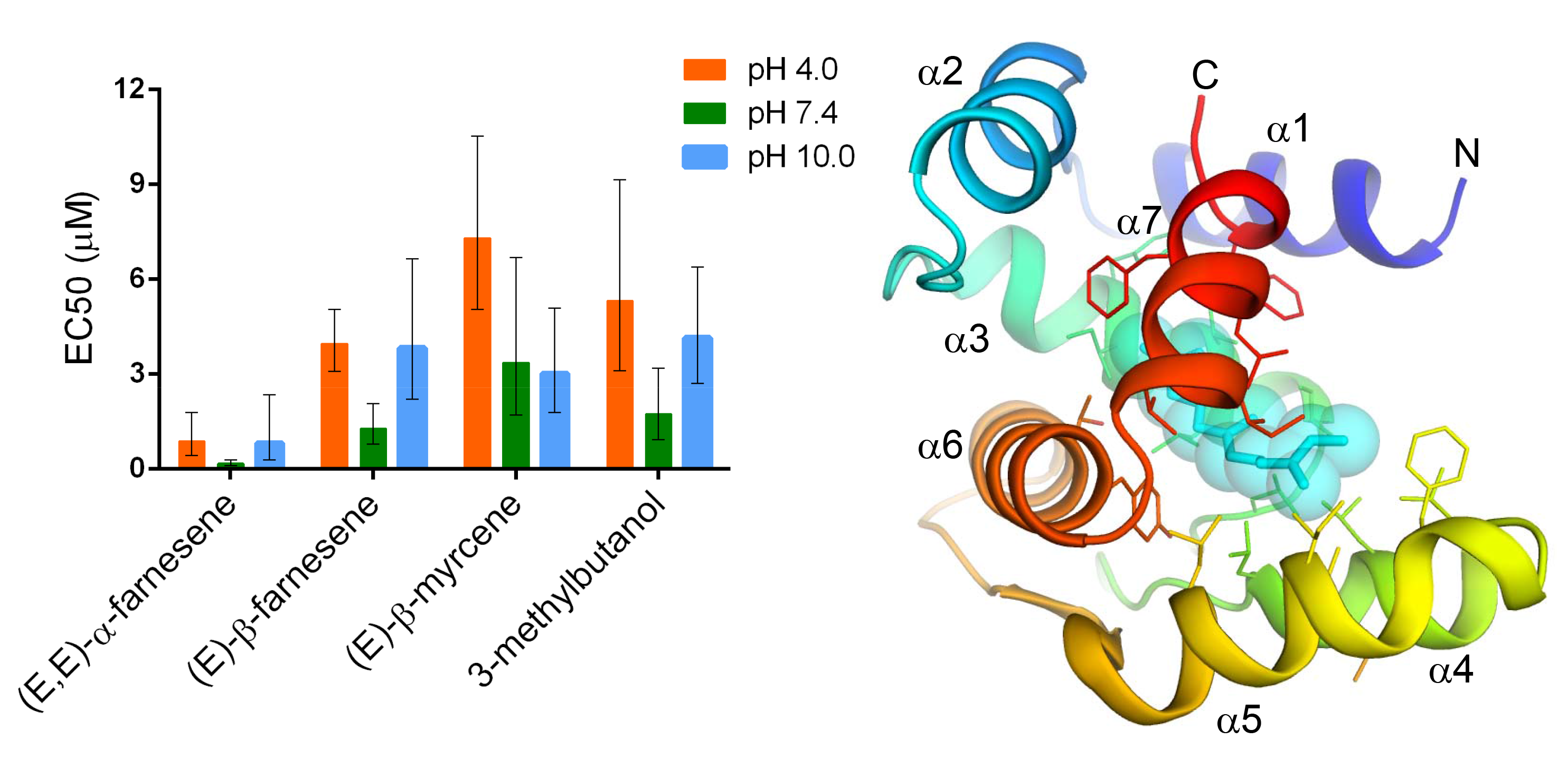
2018

Scietti L., Chiapparino A., De Giorgi F., Fumagalli M., Khoriauli L., Nergadze S., Basu S., Olieric V., Cucca L., Banushi B., Profumo A., Giulotto E., Gissen P., Forneris F.
Molecular architecture of the multifunctional collagen lysyl hydroxylase and glycosyltransferase LH3
Nature Communications, 9, 3163. (2018) - PubMed

Buroni S.*, Scoffone V.C.*, Fumagalli M., Makarov V., Cagnone M., Trespidi G., De Rossi E., Forneris F., Riccardi G., Chiarelli L.R.
Investigating the Mechanism of Action of Diketopiperazines Inhibitors of the Burkholderia cenocepacia Quorum Sensing Synthase CepI: A Site-Directed Mutagenesis Study
Frontiers in Pharmacology, 9, 836. (2018) - PubMed
*Equal Contribution 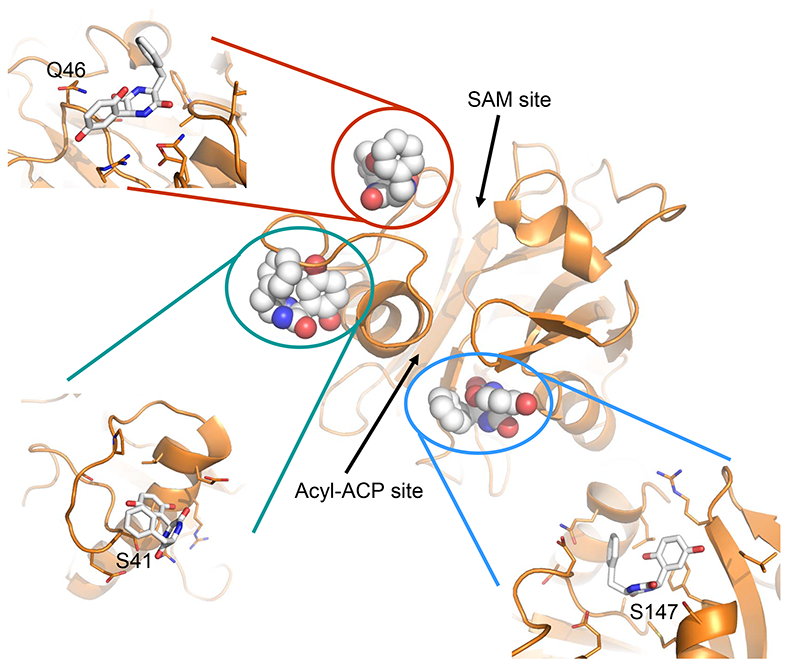
2017

Xue X., Wu J., Ricklin D., Forneris F., Di Crescenzio P., Schmidt C., Granneman J.C., Sharp T.H., Lambris J.D., Gros P.
Regulator-dependent mechanisms of C3b processing by factor I allow for differentiation of immune responses
Nature Structural and Molecular Biology, 24, 643-651. (2017) - PubMed

Forneris F.*, Mattevi A.*
Expanding the structural biology toolbox with single-molecule holography
Proceedings of the National Academy of Sciences, U.S.A., 114, 1448-1450. (2017) - PubMed
*Shared Corresponding Authors
2016

Gruber R., Rogerson C., Windpassinger C., Banushi B., Straatman-Iwanowska A., Hanley J., Forneris F., Strohal R., Ulz P., Crumrine D., Menon G.K., Blunder S., Schmuth M., Müller T., Smith H., Mills K., Kroisel P., Janecke A.R., Gissen P.
Autosomal recessive Keratoderma-Ichthyosis-Deafness (ARKID) syndrome is caused by VPS33B mutations affecting Rab protein interaction and collagen modification
Journal of Investigative Dermatology, 137, 32800-32807. (2016) - PubMed

Israyilova A., Buroni S., Forneris F., Scoffone V.C., Shixaliyev N.Q., Riccardi G., Chiarelli L.R.
Biochemical characterization of Glutamate Racemase, a new candidate drug target against Burkholderia cenocepacia infections
PLoS One, 11, e0167350. (2016) - PubMed

Speranzini V., Rotili D., Ciossani G., Pilotto S., Marrocco B., Forgione M., Lucidi A., Forneris F., Mehdipour P., Velankar S., Mai A., Mattevi A.
Polymyxins and quinazolines are LSD1/KDM1A inhibitors with unusual structural features
Science Advances, 2, e1601017. (2016) - PubMed
Watch the Movie

Scoffone V.C.*, Chiarelli L.R.*, Makarov V.*, Brackman G., Israylova A., Azzalin A., Forneris F., Riabova O., Savina S., Coenye T., Riccardi G., Buroni S.
Discovery of new diketopiperazines inhibiting Burkholderia cenocepacia quorum sensing in vitro and in vivo
Scientific Reports, 6, 32487. (2016) - PubMed
*Equal Contribution  -
- 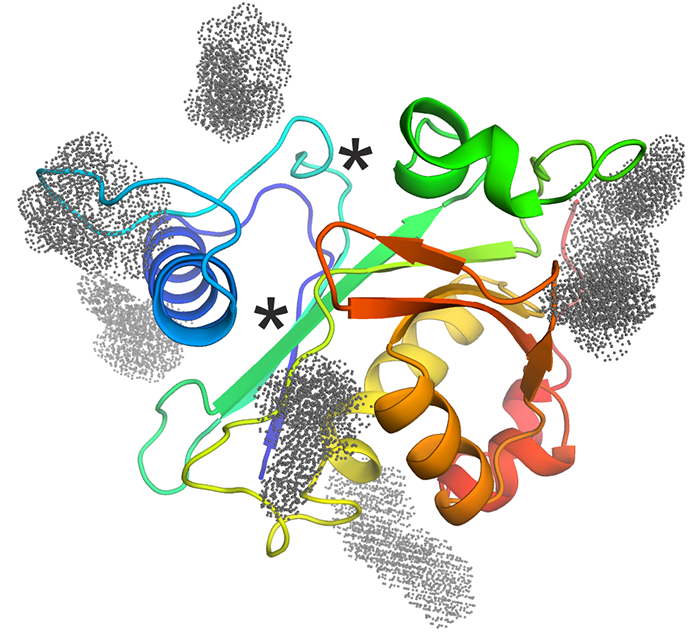

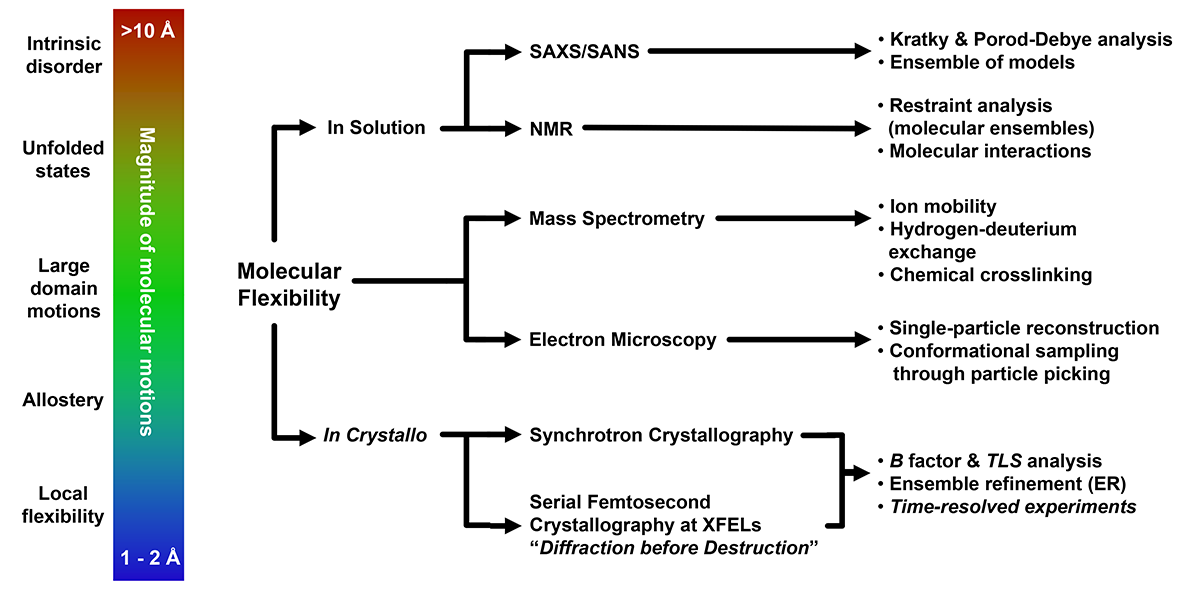
Palamini M., Canciani A., Forneris F.
Identifying and visualizing macromolecular flexibility in structural biology
Frontiers in Molecular Biosciences, 3, 47. (2016) - PubMed

Banushi B., Forneris F.*, Straatman-Iwanowska A., Strange A., Lyne A., Rogerson C., Burden J.J., Heywood W.E., Hanley J., Doykov I., Straatman K.R., Smith H., Bem D., Kriston-Vizi J., Ariceta G., Risteli M., Wang C., Ardill R.E., Zaniew M., Latka-Grot J., Waddington S.N., Howe S.J., Ferraro F., Gjinovci A., Lawrence S., Marsh M., Girolami M., Bozec L., Mills K., Gissen P.*
Regulation of post-Golgi LH3 trafficking is essential for collagen homeostasis
Nature Communications, 7, 12111. (2016) - PubMed
*Shared Corresponding Authors 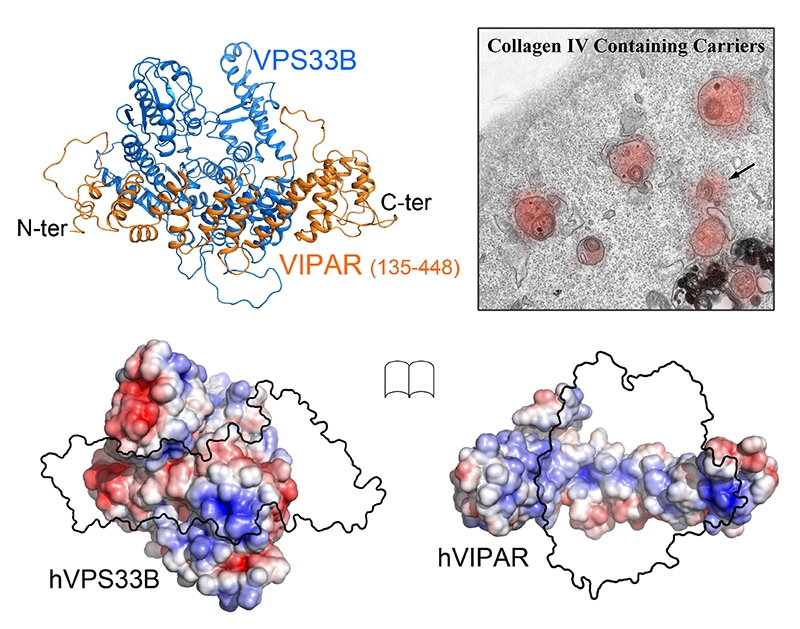
Spadaro F.*, Scoffone V.C.*, Chiarelli L.R.**, Fumagalli M., Buroni S., Riccardi G., Forneris F.**
The crystal structure of Burkholderia cenocepacia DfsA provides insights into substrate recognition and quorum sensing fatty acid biosynthesis
Biochemistry, 55, 3241-3250. (2016) - PubMed
*Equal Contribution - **Shared Corresponding Authors 

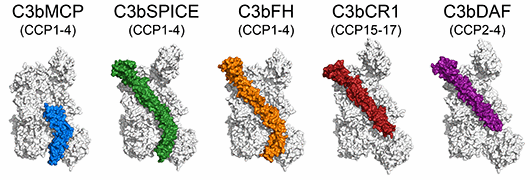
Forneris F., Wu J., Xue X., Ricklin D., Lin Z., Sfyroera G., Tzekou A., Volokhina E., Granneman J.C.M., Hauhart R., Bertram P., Liszewski M.K., Atkinson J.P., Lambris J.D., Gros P.
Regulators of complement activity mediate inhibitory mechanisms through a common C3b-binding mode
The EMBO Journal, 35, 1133-1149. (2016) - PubMed
 -
-

Savino S., Ferrandi E., Forneris F., Rovida S., Riva S., Monti D., Mattevi A.
Structural and biochemical insights into 7b-hydroxysteroid dehydrogenase stereoselectivity
Proteins, 84, 859-865. (2016) - PubMed
Cover Image 
2015

Dzurova L., Forneris F.*, Savino S., Galuszka P., Vrabka J., Frebort I.*
The three-dimensional Structure of Lonely Guy from Claviceps purpurea provides insights into the phosphoribohydrolase function of Rossmann fold-containing lysine decarboxylase-like proteins.
Proteins, 83, 1539-1546. (2015) - PubMed
*Shared Corresponding Authors
Datasets available in repository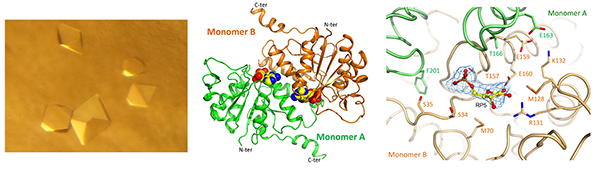
Pilotto S., Speranzini V., Tortorici M., Durand D., Fish A., Valente S., Forneris F., Mai A., Sixma, T.K., Vachette P., Mattevi A.
Interplay among nucleosomal DNA, histone tails, and corepressor CoREST underlies LSD1-mediated H3 demethylation
Proceedings of the National Academy of Sciences, U.S.A., 112, 2752-2757. (2015) - PubMed
Watch the Movie
2014

Forneris F., Burnley B.T., Gros P.
Ensemble refinement shows conformational flexibility in crystal structures of human complement factor D.
Acta Crystallographica section D - Biological Crystallography, 70, 733-743. (2014) - PubMed
2013

Peng W.C., de Lau W., Madoori P.K., Forneris F., Granneman J.C., Clevers H., Gros P.
Structures of Wnt-Antagonist ZNRF3 and Its Complex with R-Spondin 1 and Implications for Signaling.
PLoS One, 8, e83110. (2013) - PubMed

Minde D.P., Radli M., Forneris F., Maurice M.M., Rüdiger S.G.
Large extent of disorder in Adenomatous Polyposis Coli offers a strategy to guard Wnt signalling against point mutations.
PLoS One, 8, e77257. (2013) - PubMed

Peng W.C., de Lau W., Forneris F., Granneman J.C., Huch M., Clevers H., Gros P.
Structure of stem cell growth factor R-spondin 1 in complex with the ectodomain of its receptor LGR5.
Cell Reports, 3, 1885-1892. (2013) - PubMed
2012
Hadders M.A., Bubeck D., Roversi P., Hakobyan S., Forneris F., Morgan B.P., Pangburn M.K., Llorca O., Lea S.M., Gros P.
Assembly and regulation of the membrane attack complex based on structures of C5b6 and sC5b9.
Cell Reports, 1, 200-207. (2012) - PubMed
 -
-

Forneris F., Wu J., Gros P.
The modular serine proteases of the complement cascade.
Current Opinions in Structural Biology, 22, 333-341. (2012) - PubMed
2010

Forneris F., Ricklin D., Wu J., Tzekou A., Wallace R.S., Lambris J.D., Gros P.
Structures of C3b in complex with factors B and D give insight into complement convertase formation.
Science, 330, 1816-1820. (2010) - PubMed

Binda C., Valente S., Romanenghi M., Pilotto S., Cirilli R., Karytinos A., Ciossani G., Botrugno O.A., Forneris F., Tardugno M., Edmondson D.E., Minucci S., Mattevi A., Mai A.
Biochemical, structural, and biological evaluation of tranylcypromine derivatives as inhibitors of histone demethylases LSD1 and LSD2.
Journal of the American Chemical Society, 132, 6827-6833. (2010) - PubMed
Zibetti C., Adamo A., Binda C., Forneris F., Toffolo E., Verpelli C., Ginelli E., Mattevi A., Sala C., Battaglioli E.
Alternative splicing of the histone demethylase LSD1/KDM1 contributes to the modulation of neurite morphogenesis in the mammalian nervous system.
Journal of Neuroscience, 30, 2521-2532. (2010) - PubMed
2009

Villa F., Capasso P., Tortorici M., Forneris F., de Marco A., Mattevi A., Musacchio A.
Crystal structure of the catalytic domain of Haspin, an atypical kinase implicated in chromatin organization.
Proceedings of the National Academy of Sciences, U.S.A., 106, 20204-20209. (2009) - PubMed

Forneris F., Battaglioli E., Mattevi A., Binda C.
New roles of flavoproteins in molecular cell biology: histone demethylase LSD1 and chromatin.
FEBS Journal, 276, 4304-4312. (2009) - PubMed

Baron R., Riley C., Chenprakhon P., Thotsaporn K., Winter R.T., Alfieri A., Forneris F., van Berkel W.J., Chaiyen P., Fraaije M.W., Mattevi A., McCammon J.A.
Multiple pathways guide oxygen diffusion into flavoenzyme active sites.
Proceedings of the National Academy of Sciences, U.S.A., 106, 10603-10608. (2009) - PubMed

Forneris F., Orru R., Bonivento D., Chiarelli L.R., Mattevi A.
ThermoFAD, a Thermofluor-adapted flavin ad hoc detection system for protein folding and ligand binding.
FEBS Journal, 276, 2833-2840. (2009) - PubMed

Karytinos A., Forneris F., Profumo A., Ciossani G., Battaglioli E., Binda C., Mattevi A.
A novel mammalian flavin-dependent histone demethylase.
Journal of Biological Chemistry, 284, 17775-17782. (2009) - PubMed
2008

Forneris F., Mattevi A.
Enzymes without borders: mobilizing substrates, delivering products.
Science, 321, 213-216. (2008) - PubMed

Forneris F., Binda C., Battaglioli E., Mattevi A.
LSD1: oxidative chemistry for multifaceted functions in chromatin regulation.
Trends in Biochemical Sciences, 33, 181-189. (2008) - PubMed

Forneris F., Heuts D.P., Delvecchio M., Rovida S., Fraaije M.W., Mattevi A.
Structural analysis of the catalytic mechanism and stereoselectivity in Streptomyces coelicolor alditol oxidase.
Biochemistry, 47, 978-985. (2008) - PubMed
2007

Forneris F., Binda C., Adamo A., Battaglioli E., Mattevi A.
Structural basis of LSD1-CoREST selectivity in histone H3 recognition.
Journal of Biological Chemistry, 282, 20070-20074. (2007) - PubMed
 -
-
2006

Forneris F., Rovida S., Heuts D.P., Fraaije M.W., Mattevi A.
Crystallization and preliminary X-ray analysis of an alditol oxidase from Streptomyces coelicolor A3(2).
Acta Crystallographica section F - Structural Biology Communications, 62, 1298-1300. (2006) - PubMed

Forneris F., Binda C., Dall'Aglio A., Fraaije M.W., Battaglioli E., Mattevi A.
A highly specific mechanism of histone H3-K4 recognition by histone demethylase LSD1.
Journal of Biological Chemistry, 281, 35289-35295. (2006) - PubMed
2005
Forneris F., Binda C., Vanoni M.A., Battaglioli E., Mattevi A.
Human histone demethylase LSD1 reads the histone code.
Journal of Biological Chemistry, 280, 41360-41365. (2005) - PubMed

Forneris F., Binda C., Vanoni M.A., Mattevi A., Battaglioli E.
Histone demethylation catalysed by LSD1 is a flavin-dependent oxidative process.
FEBS Letters, 579, 2203-2207. (2005) - PubMed
2003

Di Costanzo L, Forneris F, Geremia S, Randaccio L.
Phasing protein structures using the group-subgroup relation.
Acta Crystallographica section D - Biological Crystallography, 59, 1435-1439. (2003) - PubMed