 About
Contact
References
Structures
Adv. Search
Stats
Demo
About
Contact
References
Structures
Adv. Search
Stats
Demo
From this page you can gather additional details and download molecular coordinates:
| Gene and cDNA details and annotations |
| NCBI Entrez Gene id: 5351 - Refseq: NM_000302.2 - Sequence viewer |
| HGNC: 9081 - ENSEMBL: ENSG00000083444 - UCSC: uc001atm.4 - GenAtlas: 557 |
| NCBI CCDS id: CCDS142 - NCBI Nuccore: NG_008159.1 |
Protein details and annotations |
| Uniprot: Q02809 - NCBI Protein: NP_000293.2 - KEGG: K00473 |
| InterPro - Pfam - Protein Sequence |
Clinical annotations |
| OMIM: 153454 - Orphanet: 117888 - LOVD: PLOD1 - DECIPHER: PLOD1 |
Available molecular coordinates for download: - homology model of full-length, dimeric human LH1 (generated using the crystal structure of full-length human LH3 as template) - Model details |
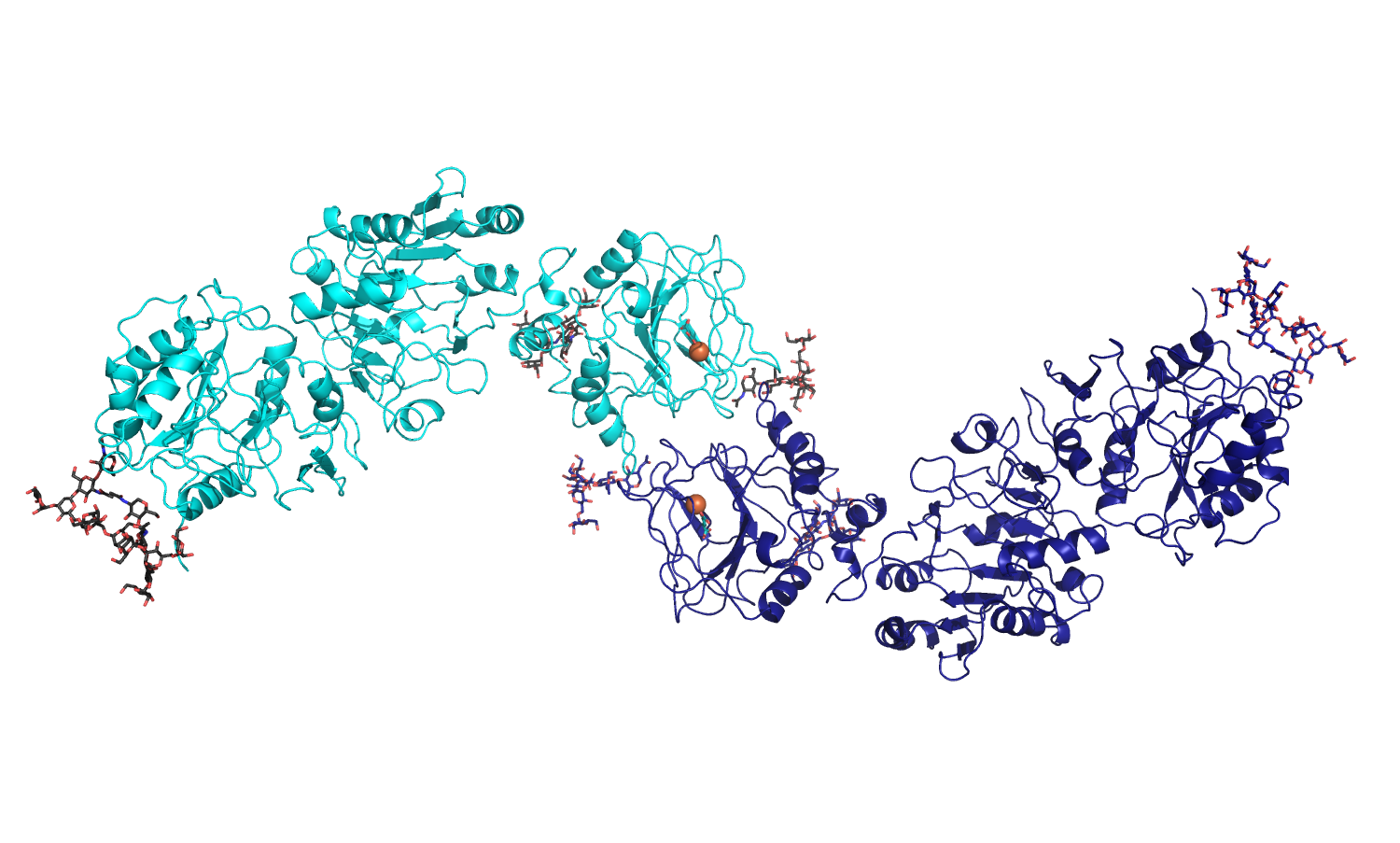
| Gene and cDNA details and annotations |
| NCBI Entrez Gene id: 5352 - Refseq: NM_000935.2 - Sequence viewer |
| HGNC: 9082 - ENSEMBL: ENSG00000152952 - UCSC: uc003evr.2 - GenAtlas: 6102 |
| NCBI CCDS id: CCDS3131 - NCBI Nuccore: NG_009251.1 |
Protein details and annotations |
| Uniprot: O00469 - NCBI Protein: NP_000926.2 - KEGG: K13645 |
| InterPro - Pfam - Protein Sequence |
Clinical annotations |
| OMIM: 601865 - Orphanet: 117892 - LOVD: PLOD2 - DECIPHER: PLOD2 |
Available molecular coordinates for download: - homology model of full-length, dimeric human LH2a (generated using the crystal structure of full-length human LH3 as template) - Model details |
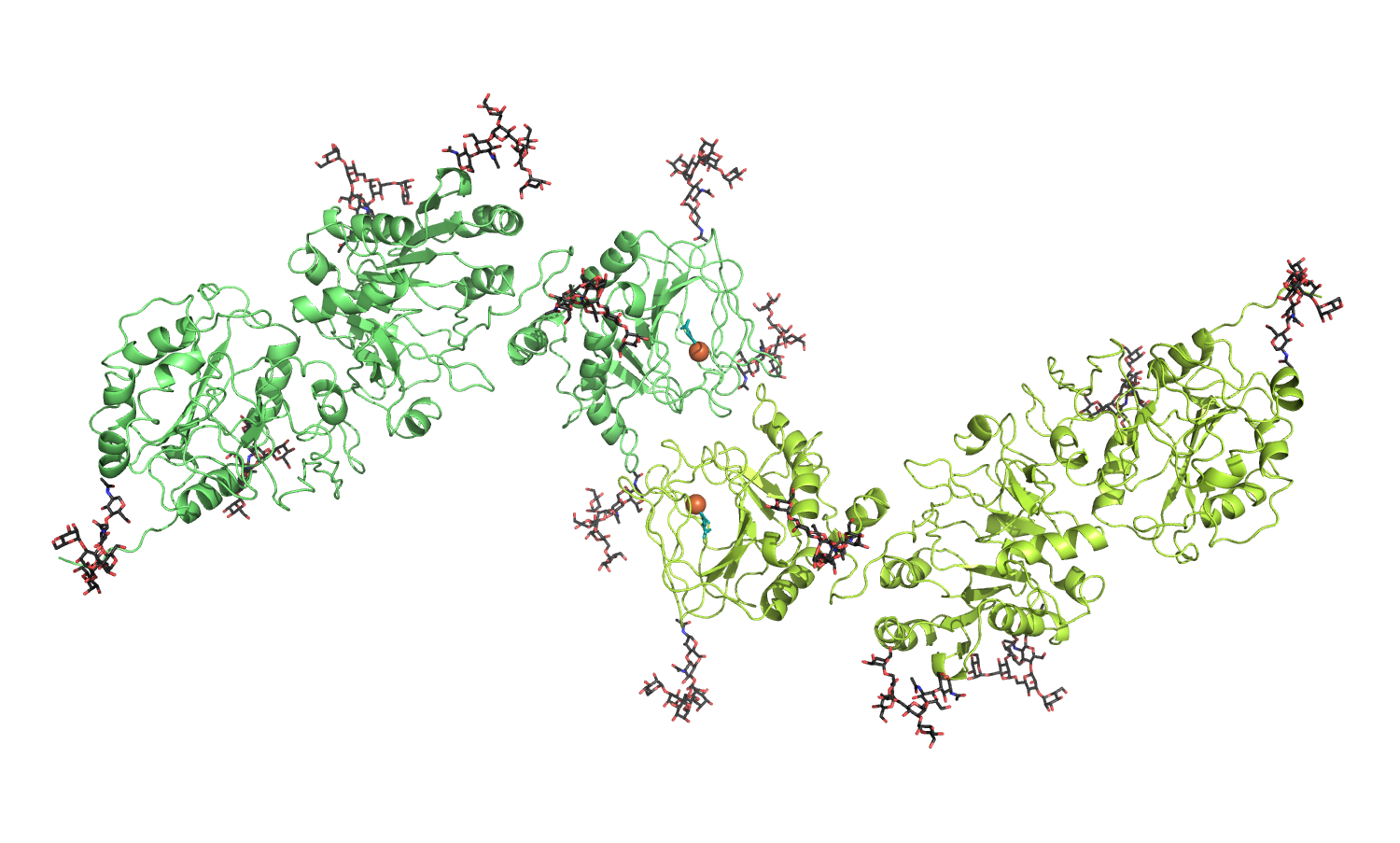
| Gene and cDNA details and annotations |
| NCBI Entrez Gene id: 5352 - Refseq: NM_182943.2 - Sequence viewer |
| HGNC: 9082 - ENSEMBL: ENSG00000152952 - UCSC: uc003evr.2 - GenAtlas: 6102 |
| NCBI CCDS id: CCDS3132 - NCBI Nuccore: NG_009251.1 |
Protein details and annotations |
| Uniprot: O00469-2 - NCBI Protein: NP_891988.1 - KEGG: K13645 |
| InterPro - Pfam - Protein Sequence |
Clinical annotations |
| OMIM: 601865 - Orphanet: 117892 - LOVD: PLOD2 - DECIPHER: PLOD2 |
Available molecular coordinates for download: - homology model of full-length, dimeric human LH2b (generated using the crystal structure of full-length human LH3 as template) - Model details |
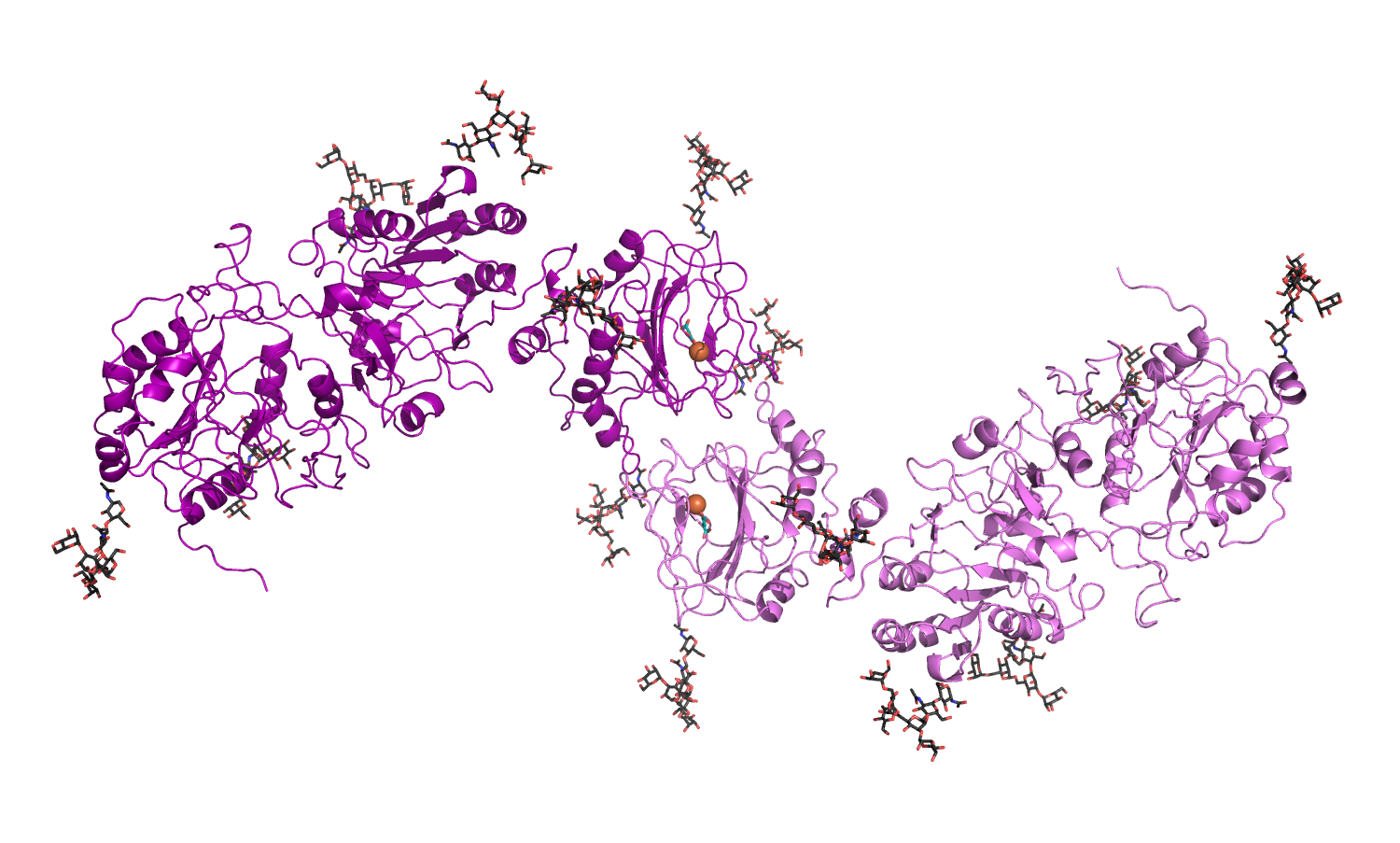
| Gene and cDNA details and annotations |
| NCBI Entrez Gene id: 8985 - Refseq: NM_001084.4 - Sequence viewer |
| HGNC: 9083 - ENSEMBL: ENSG00000106397 - UCSC: uc003uyd.4 - GenAtlas: 6873 |
| NCBI CCDS id: CCDS5715 - NCBI Nuccore: NG_012148.1 |
Protein details and annotations |
| Uniprot: O60568 - NCBI Protein: NP_001075.1 - KEGG: K13646 |
| InterPro - Pfam - Protein Sequence |
Clinical annotations |
| OMIM: 603066 - Orphanet: 304172 - LOVD: PLOD3 - DECIPHER: PLOD3 |
Available molecular coordinates for download: - homology model of full-length, dimeric human LH3 (generated using the crystal structure of full-length human LH3 as template) - Model details - dimeric structure of LH3 in complex with Fe2+ and 2-OG (from PDB id 6FXK) - dimeric structure of LH3 in complex with Fe2+, 2-OG and Mn2+ (from PDB id 6FXM) - dimeric structure of LH3 in complex with Fe2+, 2-OG, Mn2+ and UDP-Galactose (from PDB id 6FXR) - dimeric structure of LH3 in complex with Fe2+, 2-OG, Mn2+ and UDP-Glucose (from PDB id 6FXT) |
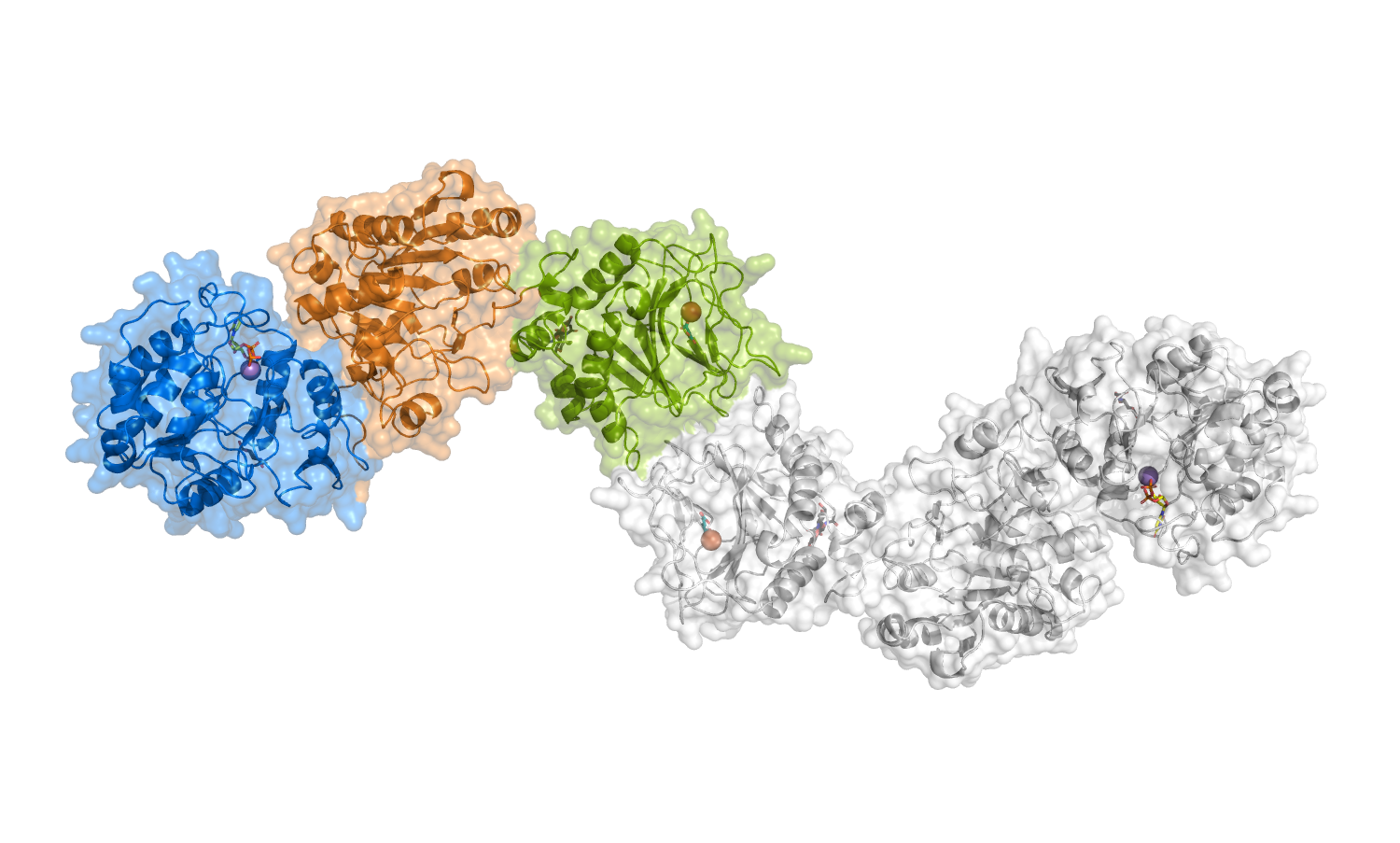
| Gene and cDNA details and annotations |
| NCBI Entrez Gene id: 9924837 - Refseq: HQ336222.2 - Sequence viewer |
Protein details and annotations |
| Uniprot: Q5UQC3 - NCBI Protein: YP_003986726.1 - KEGG: K21236 |
| InterPro - Pfam - Protein Sequence |
Available molecular coordinates for download: - dimeric structure of of the C-terminal LH domain of mimivirus L230 in complex with Fe2+ (from PDB id 6AX7) |
Thank you for using SiMPLOD - Created by Fornerislab@UniPV Follow @Fornerislab - Last curated update: 1970-01-01 00:00:00
We truly hate messages and disclaimers about cookies and tracking of personal info. But don't worry, we don't use any.