 About
Contact
References
Structures
Adv. Search
Stats
Demo
About
Contact
References
Structures
Adv. Search
Stats
Demo
a Structurally-integrated database for Mutations of PLOD genes
SiMPLOD is a manually-curated PHP-mySQL online database which collects all the currently described natural variants as well as biochemical mutations of PLOD enzymes and maps them using
available molecular structure information.
Mutations in the LH/PLOD (Procollagen-Lysine 2-Oxoglutarate 5-Dioxygenase/Lysyl-Hydroxylase) enzyme family (constituted by the three isoforms LH1/PLOD1, LH2/PLOD2 and LH3/PLOD3) cause severe connective tissue disorders generally characterized by bone fragility, contractures of variable severity and different skeletal manifestations. These include the kyphoscholiotic and cardiovascular manifestations of Ehlers-Danlos (type VIa) syndrome, Bruck syndrome, and developmental disorders with phenotypes resembling osteogenesis imperfecta. The comprehensive biochemical and medical knowledge accumulated over more than forty years can nowadays benefit of inferring molecular structure information.
Search
for genetic variants reported in the human PLOD1, PLOD2, and PLOD3 genes to obtain structural mapping of these mutations on
gene products, either using experimentally determined crystal structures (for LH3/PLOD3) or using computational homology models of LH1/PLOD1 and LH2/PLOD2 obtained using the LH3/PLOD3 crystal structures as templates. For each variant, the change in nucleotide sequence and the corresponding change in aminoacid sequence is shown. The disease phenotype is reported for pathogenic mutations. Each mutation page, is accompanied by additional information from literature (main citation, available information on enzymatic activity), annotations about its localization on the enzyme's three-dimensional structure, and on the mutation and useful links to the main mutation databases
(NCBI gene, Uniprot, NCBI SNP)
, as well as disease phenotype information and specific links to disease annotation databases
(OMIM, Orphanet, ICD-10, MeSH)
in the case of pathogenic mutations.
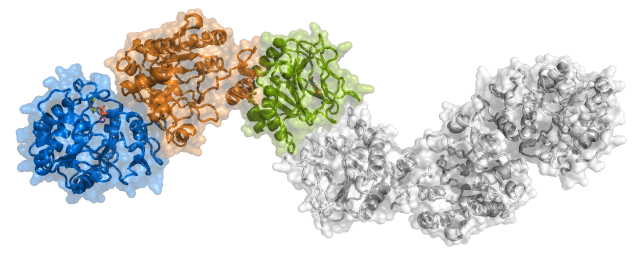
The SiMPLOD interface allows localization of the desired mutation(s) on the protein structure, enabling structure browsing and generation of high-definition structural visualizations also for non non-expert users. Several options for structure representation and colouring are present, as well as options for mutations and domain and mutation centering. As default option, the LH/PLOD dimer is coloured using blue for the N-terminal glycosyltranferase (GT) domain, orange for the central accessory (AC) domain and green for the C-terminal lysyl-hydroxylase (LH) domain. For mutations generating premature termination codon, the truncated portion of the enzyme is coloured in white, and disclaimers about possible unfolding/complete lack of expression are present. A drop down menu allows to select the molecular structure for visualization. You can test the viewer using
the demo page, which randomly picks a mutation from the database.
Thank you for using SiMPLOD - Created by Fornerislab@UniPV Follow @Fornerislab - Last curated update: 1970-01-01 00:00:00
We truly hate messages and disclaimers about cookies and tracking of personal info. But don't worry, we don't use any.